Characterization of Microsatellite Loci Markers for the Critically Endangered Montseny Brook Newt (Calotriton Arnoldi)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

What Really Hampers Taxonomy and Conservation? a Riposte to Garnett and Christidis (2017)
Zootaxa 4317 (1): 179–184 ISSN 1175-5326 (print edition) http://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2017 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4317.1.10 http://zoobank.org/urn:lsid:zoobank.org:pub:88FA0944-D3CF-4A7D-B8FB-BAA6A3A76744 What really hampers taxonomy and conservation? A riposte to Garnett and Christidis (2017) MARCOS A. RAPOSO1,2, RENATA STOPIGLIA3,4, GUILHERME RENZO R. BRITO1,5, FLÁVIO A. BOCKMANN3,6, GUY M. KIRWAN1,7, JEAN GAYON2 & ALAIN DUBOIS4 1 Setor de Ornitologia, Departamento de Vertebrados, Museu Nacional, Universidade Federal do Rio de Janeiro, Quinta da Boa Vista, s/n, 20940–040 Rio de Janeiro, RJ, Brazil. [email protected] (MAR), [email protected] (GRRB), [email protected] (GMK) 2 UMR 8590, IHPST–Institut d'Histoire et de Philosophie des Sciences et des Techniques, UMR 8590, Université Paris 1 Panthéon- Sorbonne & CNRS, 13 rue du Four, 75006 Paris, France. [email protected] (JG) 3 Laboratório de Ictiologia de Ribeirão Preto, Departamento de Biologia, FFCLRP, Universidade de São Paulo, Av. dos Bandeirantes 3900, 14040–901 Ribeirão Preto, SP, Brazil. [email protected] (RS), [email protected] (FAB) 4 Institut de Systématique, Évolution, Biodiversité, ISYEB – UMR 7205 – CNRS, MNHN, UPMC, EPHE, Muséum national d’Histoire naturelle, Sorbonne Universités, 25 rue Cuvier, CP 30, 75005, Paris, France. [email protected] (AD). 5 Departamento de Zoologia, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Av. Carlos Chagas Filho, 373, 21941- 902, Rio de Janeiro, RJ, Brazil. 6 Programa de Pós-Graduação em Biologia Comparada, FFCLRP, Universidade de São Paulo, Av. -

Notophthalmus Perstriatus) Version 1.0
Species Status Assessment for the Striped Newt (Notophthalmus perstriatus) Version 1.0 Striped newt eft. Photo credit Ryan Means (used with permission). May 2018 U.S. Fish and Wildlife Service Region 4 Jacksonville, Florida 1 Acknowledgements This document was prepared by the U.S. Fish and Wildlife Service’s North Florida Field Office with assistance from the Georgia Field Office, and the striped newt Species Status Assessment Team (Sabrina West (USFWS-Region 8), Kaye London (USFWS-Region 4) Christopher Coppola (USFWS-Region 4), and Lourdes Mena (USFWS-Region 4)). Additionally, valuable peer reviews of a draft of this document were provided by Lora Smith (Jones Ecological Research Center) , Dirk Stevenson (Altamaha Consulting), Dr. Eric Hoffman (University of Central Florida), Dr. Susan Walls (USGS), and other partners, including members of the Striped Newt Working Group. We appreciate their comments, which resulted in a more robust status assessment and final report. EXECUTIVE SUMMARY This Species Status Assessment (SSA) is an in-depth review of the striped newt's (Notophthalmus perstriatus) biology and threats, an evaluation of its biological status, and an assessment of the resources and conditions needed to maintain species viability. We begin the SSA with an understanding of the species’ unique life history, and from that we evaluate the biological requirements of individuals, populations, and species using the principles of population resiliency, species redundancy, and species representation. All three concepts (or analogous ones) apply at both the population and species levels, and are explained that way below for simplicity and clarity as we introduce them. The striped newt is a small salamander that uses ephemeral wetlands and the upland habitat (scrub, mesic flatwoods, and sandhills) that surrounds those wetlands. -

Calotriton Asper)
Amphibia-Reptilia (2014) DOI:10.1163/15685381-00002921 Life history trait differences between a lake and a stream-dwelling population of the Pyrenean brook newt (Calotriton asper) Neus Oromi1,∗, Fèlix Amat2, Delfi Sanuy1, Salvador Carranza3 Abstract. The Pyrenean brook newt (Calotriton asper) is a salamandrid that mostly lives in fast running and cold mountain- streams, although some populations are also found in lakes. In the present work, we report in detail on the occurrence of facultative paedomorphosis traits in a population from a Pyrenean high altitude lake. We compare its morphology, life history traits and mitochondrial DNA variation with a nearby lotic metamorphic population. Our results indicate that the lacustrine newts are smaller and present a less developed sexual dimorphism, smooth skin, and that 53% of the adults retain gills at different degrees of development, but not gill slits. Although both populations and sexes have the same age at sexual maturity (four years), the lacustrine population presents higher longevity (12 and 9 years for males and females, respectively) than the one living in the stream (8 and 9 years). The variation on the climatic conditions at altitudinal scale is probably the main cause of the differences in life history traits found between the two populations. The food availability, which could to be limiting in the lacustrine population, is another factor that can potentially affect body size. These results are congruent with the significant mitochondrial DNA genetic isolation between populations, probably a consequence of the lack of juvenile dispersal. We found low cytochrome b variability and significant genetic structuring in the lake population that is very remarkably considering the short distance to the nearby stream population and the whole species’ pattern. -
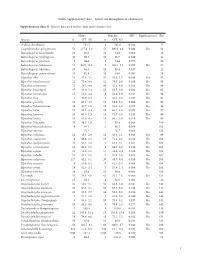
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -

Unraveling the Rapid Radiation of Crested Newts (Triturus Cristatus Superspecies) Using Complete Mitogenomic Sequences Ben Wielstra1,2* and Jan W Arntzen2
Wielstra and Arntzen BMC Evolutionary Biology 2011, 11:162 http://www.biomedcentral.com/1471-2148/11/162 RESEARCH ARTICLE Open Access Unraveling the rapid radiation of crested newts (Triturus cristatus superspecies) using complete mitogenomic sequences Ben Wielstra1,2* and Jan W Arntzen2 Abstract Background: The rapid radiation of crested newts (Triturus cristatus superspecies) comprises four morphotypes: 1) the T. karelinii group, 2) T. carnifex - T. macedonicus,3)T. cristatus and 4) T. dobrogicus. These vary in body build and the number of rib-bearing pre-sacral vertebrae (NRBV). The phylogenetic relationships of the morphotypes have not yet been settled, despite several previous attempts, employing a variety of molecular markers. We here resolve the crested newt phylogeny by using complete mitochondrial genome sequences. Results: Bayesian inference based on the mitogenomic data yields a fully bifurcating, significantly supported tree, though Maximum Likelihood inference yields low support values. The internal branches connecting the morphotypes are short relative to the terminal branches. Seen from the root of Triturus (NRBV = 13), a basal dichotomy separates the T. karelinii group (NRBV = 13) from the remaining crested newts. The next split divides the latter assortment into T. carnifex - T. macedonicus (NRBV = 14) versus T. cristatus (NRBV = 15) and T. dobrogicus (NRBV = 16 or 17). Conclusions: We argue that the Bayesian full mitochondrial DNA phylogeny is superior to previous attempts aiming to recover the crested newt species tree. Furthermore, our new phylogeny involves a maximally parsimonious interpretation of NRBV evolution. Calibrating the phylogeny allows us to evaluate potential drivers for crested newt cladogenesis. The split between the T. -

Incidence of Emerging Pathogens in the Legal and Illegal Amphibian Trade in Spain
Herpetology Notes, volume 14: 777-784 (2021) (published online on 16 May 2021) Incidence of emerging pathogens in the legal and illegal amphibian trade in Spain Barbora Thumsová1,2, Jaime Bosch3,2,*, and Albert Martinez-Silvestre4 Abstract. Amphibians are threatened globally and emerging diseases are some of the most important drivers of their catastrophic situation. There is increasing evidence that the international trade in live amphibians is one of the most important mechanisms driving pathogen pollution. Here, we report the presence of Batrachochytrium dendrobatidis (Bd) and Ranavirus in 11% of tested individuals in legal amphibian trade fairs in Spain. Although none of the Bd infected animals in trade fairs presented disease symptoms, symptoms of ranavirosis were observed in some specimens, which were nonetheless still offered for sale. None of the traders who were selling infected animals showed interest in engaging in collaboration to control infections when offered for free. In addition, a large private urodele collection confiscated by the police in Barcelona comprised a number of illegally wild-caught species. Many confiscated individuals presented signs of poor welfare and several were positive for Bd. Our results indicate the urgent need for implementation of real sanitary regulations or effective legislation governing the practice of trade in living amphibians to prevent pathogen spread in Europe. Keywords. Batrachochytridium dendrobatidis, chytridiomycosis, Ranavirus, ranavirosis, pet keeping Introduction al., 2005; Collins and Crump, 2009; Rowley et al., 2010; Pasmans et al., 2017). In fact, of the 5,579 vertebrate Wildlife continues to be threatened by a number of species affected by the global wildlife trade, 9.4% are anthropogenic activities that have compromised the amphibians (Scheffers et al., 2019). -

Amazing Amphibians Celebrating a Decade of Amphibian Conservation
QUARTERLY PUBLICATION OF THE EUROPEAN ASSOCIATION OF ZOOS AND AQUARIA AUTUMNZ 2018OO QUARIAISSUE 102 AMAZING AMPHIBIANS CELEBRATING A DECADE OF AMPHIBIAN CONSERVATION A giant challenge BUILDING A FUTURE FOR THE CHINESE GIANT SALAMANDER 1 Taking a Leap PROTECTING DARWIN’S FROG IN CHILE Give your visitors a digital experience Add a new dimension to your visitor experience with the Aratag app – for museums, parks and tourist www.aratag.com attractions of all kinds. Aratag is a fully-integrated information system featuring a CMS and universal app that visitors download to their smart devices. The app runs automatically when it detects a nearby facility using the Aratag system. With the power of Aratag’s underlying client CMS system, zoos, aquariums, museums and other tourist attractions can craft customized, site-specifi c app content for their visitors. Aratag’s CMS software makes it easy for you to create and update customized app content, including menus, text, videos, AR, and active links. Aratag gives you the power to intelligently monitor visitors, including demographics and visitor fl ows, visit durations, preferred attractions, and more. You can also send push messages through the app, giving your visitors valuable information such as feeding times, closing time notices, transport information, fi re alarms, evacuation routes, lost and found, etc. Contact Pangea Rocks for an on-site demonstration of how Aratag gives you the power to deliver enhanced visitor experiences. Contact us for more information: Address: Aratag is designed and Email: [email protected] Aratag / Pangea Rocks A/S developed by Pangea Rocks A/S Phone: +45 60 94 34 32 Navervej 13 in collaboration with Aalborg Mobile : +45 53 80 34 32 6800 Varde, Denmark University. -

Volume 2, Chapter 14-8: Salamander Mossy Habitats
Glime, J. M. and Boelema, W. J. 2017. Salamander Mossy Habitats. Chapt. 14-8. In: Glime, J. M. Bryophyte Ecology. Volume 2. 14-8-1 Bryological Interaction.Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 19 July 2020 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology2/>. CHAPTER 14-8 SALAMANDER MOSSY HABITATS Janice M. Glime and William J. Boelema TABLE OF CONTENTS Tropical Mossy Habitats – Plethodontidae........................................................................................................ 14-8-3 Terrestrial and Arboreal Adaptations ......................................................................................................... 14-8-3 Bolitoglossa (Tropical Climbing Salamanders) ......................................................................................... 14-8-4 Bolitoglossa diaphora ................................................................................................................................ 14-8-5 Bolitoglossa diminuta (Quebrada Valverde Salamander) .......................................................................... 14-8-5 Bolitoglossa hartwegi (Hartweg's Mushroomtongue Salamander) ............................................................ 14-8-5 Bolitoglossa helmrichi ............................................................................................................................... 14-8-5 Bolitoglossa jugivagans ............................................................................................................................ -

July to December 2019 (Pdf)
2019 Journal Publications July Adelizzi, R. Portmann, J. van Meter, R. (2019). Effect of Individual and Combined Treatments of Pesticide, Fertilizer, and Salt on Growth and Corticosterone Levels of Larval Southern Leopard Frogs (Lithobates sphenocephala). Archives of Environmental Contamination and Toxicology, 77(1), pp.29-39. https://www.ncbi.nlm.nih.gov/pubmed/31020372 Albecker, M. A. McCoy, M. W. (2019). Local adaptation for enhanced salt tolerance reduces non‐ adaptive plasticity caused by osmotic stress. Evolution, Early View. https://onlinelibrary.wiley.com/doi/abs/10.1111/evo.13798 Alvarez, M. D. V. Fernandez, C. Cove, M. V. (2019). Assessing the role of habitat and species interactions in the population decline and detection bias of Neotropical leaf litter frogs in and around La Selva Biological Station, Costa Rica. Neotropical Biology and Conservation 14(2), pp.143– 156, e37526. https://neotropical.pensoft.net/article/37526/list/11/ Amat, F. Rivera, X. Romano, A. Sotgiu, G. (2019). Sexual dimorphism in the endemic Sardinian cave salamander (Atylodes genei). Folia Zoologica, 68(2), p.61-65. https://bioone.org/journals/Folia-Zoologica/volume-68/issue-2/fozo.047.2019/Sexual-dimorphism- in-the-endemic-Sardinian-cave-salamander-Atylodes-genei/10.25225/fozo.047.2019.short Amézquita, A, Suárez, G. Palacios-Rodríguez, P. Beltrán, I. Rodríguez, C. Barrientos, L. S. Daza, J. M. Mazariegos, L. (2019). A new species of Pristimantis (Anura: Craugastoridae) from the cloud forests of Colombian western Andes. Zootaxa, 4648(3). https://www.biotaxa.org/Zootaxa/article/view/zootaxa.4648.3.8 Arrivillaga, C. Oakley, J. Ebiner, S. (2019). Predation of Scinax ruber (Anura: Hylidae) tadpoles by a fishing spider of the genus Thaumisia (Araneae: Pisauridae) in south-east Peru. -

The Phylogeny of Crested Newts (Triturus Cristatus Superspecies)
Contributions to Zoology, 76 (4) 261-278 (2007) The phylogeny of crested newts (Triturus cristatus superspecies): nuclear and mitochondrial genetic characters suggest a hard polytomy, in line with the paleogeography of the centre of origin J.W. Arntzen1, G. Espregueira Themudo1,2, B. Wielstra1,3 1 National Museum of Natural History, P. O. Box 9517, 2300 RA Leiden, The Netherlands, [email protected] 2 CIBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos, Campus Agrário de Vairão, 4485-661 Vairão, Portugal, [email protected] 3 Institute of Biology Leiden, Leiden University, P.O. Box 9516, 2300 RA Leiden, The Netherlands, wielstra@natu- ralis.nl Key words: allozymes, historical biogeography, mitochondrial DNA-sequences, Triturus macedonicus, Triturus marmoratus, vicariance Abstract tionary lineage and we propose to elevate its taxonomic status to that of a species, i.e., from Triturus c. macedonicus (Karaman, Newts of the genus Triturus (Amphibia, Caudata, Salamandri- 1922) to Triturus macedonicus (Karaman, 1922). dae) are distributed across Europe and adjacent Asia. In spite of its prominence as a model system for evolutionary research, the phylogeny of Triturus has remained incompletely solved. Our Contents aim was to rectify this situation, to which we employed nuclear encoded proteins (40 loci) and mitochondrial DNA-sequence Introduction .................................................................................... 261 data (mtDNA, 642 bp of the ND4 gene). We sampled up to four Materials and methods ................................................................. 263 populations per species covering large parts of their ranges. Al- Sampling strategy ................................................................... 263 lozyme and mtDNA data were analyzed separately with parsi- Allozyme data .......................................................................... 263 mony, distance, likelihood and Bayesian methods of phyloge- Mitochondrial DNA-sequence data ................................... -

Calotriton Arnoldi Carranza and Amat, 2005 Class: Amphibia Order: Caudata Family: Salamandridae Genus: Calotriton
Daniel Hernández Alonso Introduction: Taxonomy Description Distribution Habitat Ecology Value Status review: Conservation status, Populations trend Photo: Felix Amat during Threat analysis species monitoring Conservation strategies: Conservation actions in place, conservation actions needed and research needed Life Tritó Montseny Objectives Actions Problems References Introduction Taxonomy: Calotriton arnoldi Carranza and Amat, 2005 Class: Amphibia Order: Caudata Family: Salamandridae Genus: Calotriton Description: Medium size (< 12 cm) – Average: 9,61cm males / 10 cm females (Carranza and Amat, 2005) Flattened head, elongated body, and tail compressed laterally (shorter and wider in males) Rough skin (Less than Calotriton asper (Dugès, 1852)) Color: Brown in dorsal side, cream color translucent in the ventral area Distribution Montseny massif, Montseny Natural Park – 10km2 2 separated “populations” without genetical flow between them IUCN (International Union for Conservation of Nature), Conservation International & NatureServe. Calotriton arnoldi. The IUCN Red List of Threatened Species. IUCN 2009 Habitat Habitat: Water streams [none individual has been seen in land yet (Montori and Campeny, 1991; Carranza and Amat, 2005)] . Strong flow of water (oxygenated) and low temperature (below 15 oC). Altitude range: 600 – 1200m Vegetation: Preference Beech forest (Fagus sylvatica L.), also in oak forest (Quercus ilex L.), or shores with salows (Salix atrocinerea Brot.) or alders (Alnus glutinosa (L.)Gaertn.), moss and ferns. Photo: Felix Amat during species monitoring Ecology Ecology: not much information about it due to its recent separation as a species Elusive species: Nocturnal and fissuring habits No data of predators Diet: Almost unknown. Predation on salamander larvae and it has seen looking for invertebrates between the rocks. Anti-predatory strategies: segregates a sticky and smelly mucous substance in the dorsal area Reproductive behaviour: still to be studied, amplexus similar to C.asper. -

Triturus Cristatus Superspecies) Using Complete Mitogenomic Sequences Ben Wielstra1,2* and Jan W Arntzen2
Wielstra and Arntzen BMC Evolutionary Biology 2011, 11:162 http://www.biomedcentral.com/1471-2148/11/162 RESEARCH ARTICLE Open Access Unraveling the rapid radiation of crested newts (Triturus cristatus superspecies) using complete mitogenomic sequences Ben Wielstra1,2* and Jan W Arntzen2 Abstract Background: The rapid radiation of crested newts (Triturus cristatus superspecies) comprises four morphotypes: 1) the T. karelinii group, 2) T. carnifex - T. macedonicus,3)T. cristatus and 4) T. dobrogicus. These vary in body build and the number of rib-bearing pre-sacral vertebrae (NRBV). The phylogenetic relationships of the morphotypes have not yet been settled, despite several previous attempts, employing a variety of molecular markers. We here resolve the crested newt phylogeny by using complete mitochondrial genome sequences. Results: Bayesian inference based on the mitogenomic data yields a fully bifurcating, significantly supported tree, though Maximum Likelihood inference yields low support values. The internal branches connecting the morphotypes are short relative to the terminal branches. Seen from the root of Triturus (NRBV = 13), a basal dichotomy separates the T. karelinii group (NRBV = 13) from the remaining crested newts. The next split divides the latter assortment into T. carnifex - T. macedonicus (NRBV = 14) versus T. cristatus (NRBV = 15) and T. dobrogicus (NRBV = 16 or 17). Conclusions: We argue that the Bayesian full mitochondrial DNA phylogeny is superior to previous attempts aiming to recover the crested newt species tree. Furthermore, our new phylogeny involves a maximally parsimonious interpretation of NRBV evolution. Calibrating the phylogeny allows us to evaluate potential drivers for crested newt cladogenesis. The split between the T.