Deep Image Prior for Undersampling High-Speed Photoacoustic Microscopy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NTGAN: Learning Blind Image Denoising Without Clean Reference
RUI ZHAO, DANIEL P.K. LUN, KIN-MAN LAM: NTGAN 1 NTGAN: Learning Blind Image Denoising without Clean Reference Rui Zhao Department of Electronic and [email protected] Information Engineering, Daniel P.K. Lun The Hong Kong Polytechnic University, [email protected] Hong Kong, China Kin-Man Lam [email protected] Abstract Recent studies on learning-based image denoising have achieved promising perfor- mance on various noise reduction tasks. Most of these deep denoisers are trained ei- ther under the supervision of clean references, or unsupervised on synthetic noise. The assumption with the synthetic noise leads to poor generalization when facing real pho- tographs. To address this issue, we propose a novel deep unsupervised image-denoising method by regarding the noise reduction task as a special case of the noise transference task. Learning noise transference enables the network to acquire the denoising ability by only observing the corrupted samples. The results on real-world denoising benchmarks demonstrate that our proposed method achieves state-of-the-art performance on remov- ing realistic noises, making it a potential solution to practical noise reduction problems. 1 Introduction Convolutional neural networks (CNNs) have been widely studied over the past decade in various computer vision applications, including image classification [13], object detection [29], and image restoration [35]. In spite of high effectiveness, CNN models always require a large-scale dataset with reliable reference for training. However, in terms of low-level vision tasks, such as image denoising and image super-resolution, the reliable reference data is generally hard to access, which makes these tasks more challenging and difficult. -

Designing Commutative Cascades of Multidimensional Upsamplers And
IEEE SIGNAL PROCESSING LETTERS: SPL.SP.4.1 THEORY, ALGORITHMS, AND SYSTEMS 0 Designing Commutative Cascades of Multidimensional Upsamplers and Downsamplers Brian L. Evans, Member, IEEE Abstract In multiple dimensions, the cascade of an upsampler by L and a downsampler by L commutes if and only if the integer matrices L and M are right coprime and LM = ML. This pap er presents algorithms to design L and M that yield commutative upsampler/dowsampler cascades. We prove that commutativity is p ossible if the 1 Jordan canonical form of the rational resampling matrix R = LM is equivalent to the Smith-McMillan form of R. A necessary condition for this equivalence is that R has an eigendecomp osition and the eigenvalues are rational. B. L. Evans is with the Department of Electrical and Computer Engineering, The UniversityofTexas at Austin, Austin, TX 78712-1084, USA. E-mail: [email protected], Web: http://www.ece.utexas.edu/~b evans, Phone: 512 232-1457, Fax: 512 471-5907. This work was sp onsored in part by NSF CAREER Award under Grant MIP-9702707. July 31, 1997 DRAFT IEEE SIGNAL PROCESSING LETTERS: SPL.SP.4.1 THEORY, ALGORITHMS, AND SYSTEMS 1 I. Introduction 1 Resampling systems scale the sampling rate by a rational factor R = L=M = LM , or 1 equivalently decimate by H = M=L = L M [1], by essentially upsampling by L, ltering, and downsampling by M . In converting compact disc data sampled at 44.1 kHz to digital audio tap e 48000 Hz 160 data sampled at 48 kHz, R = = . Because we can always factor R into coprime 44100 Hz 147 integers L and M , we can always commute the upsampler and downsampler which leads to ecient p olyphase structures of the resampling system. -

High-Quality Self-Supervised Deep Image Denoising
High-Quality Self-Supervised Deep Image Denoising Samuli Laine Tero Karras Jaakko Lehtinen Timo Aila NVIDIA∗ NVIDIA NVIDIA, Aalto University NVIDIA Abstract We describe a novel method for training high-quality image denoising models based on unorganized collections of corrupted images. The training does not need access to clean reference images, or explicit pairs of corrupted images, and can thus be applied in situations where such data is unacceptably expensive or im- possible to acquire. We build on a recent technique that removes the need for reference data by employing networks with a “blind spot” in the receptive field, and significantly improve two key aspects: image quality and training efficiency. Our result quality is on par with state-of-the-art neural network denoisers in the case of i.i.d. additive Gaussian noise, and not far behind with Poisson and impulse noise. We also successfully handle cases where parameters of the noise model are variable and/or unknown in both training and evaluation data. 1 Introduction Denoising, the removal of noise from images, is a major application of deep learning. Several architectures have been proposed for general-purpose image restoration tasks, e.g., U-Nets [23], hierarchical residual networks [20], and residual dense networks [31]. Traditionally, the models are trained in a supervised fashion with corrupted images as inputs and clean images as targets, so that the network learns to remove the corruption. Lehtinen et al. [17] introduced NOISE2NOISE training, where pairs of corrupted images are used as training data. They observe that when certain statistical conditions are met, a network faced with the impossible task of mapping corrupted images to corrupted images learns, loosely speaking, to output the “average” image. -

Learning Deep Image Priors for Blind Image Denoising
Learning Deep Image Priors for Blind Image Denoising Xianxu Hou 1 Hongming Luo 1 Jingxin Liu 1 Bolei Xu 1 Ke Sun 2 Yuanhao Gong 1 Bozhi Liu 1 Guoping Qiu 1,3 1 College of Information Engineering and Guangdong Key Lab for Intelligent Information Processing, Shenzhen University, China 2 School of Computer Science, The University of Nottingham Ningbo China 3 School of Computer Science, The University of Nottingham, UK Abstract ods comes from the effective image prior modeling over the input images [5, 14, 20]. State of the art model-based meth- Image denoising is the process of removing noise from ods such as BM3D [12] and WNNM [20] can be further noisy images, which is an image domain transferring task, extended to remove unknown noises. However, there are a i.e., from a single or several noise level domains to a photo- few drawbacks of these methods. First, these methods usu- realistic domain. In this paper, we propose an effective im- ally involve a complex and time-consuming optimization age denoising method by learning two image priors from process in the testing stage. Second, the image priors em- the perspective of domain alignment. We tackle the domain ployed in most of these approaches are hand-crafted, such alignment on two levels. 1) the feature-level prior is to learn as nonlocal self-similarity and gradients, which are mainly domain-invariant features for corrupted images with differ- based on the internal information of the input image without ent level noise; 2) the pixel-level prior is used to push the any external information. -

Layered Neural Rendering for Retiming People in Video
Layered Neural Rendering for Retiming People in Video ERIKA LU, University of Oxford, Google Research FORRESTER COLE, Google Research TALI DEKEL, Google Research WEIDI XIE, University of Oxford ANDREW ZISSERMAN, University of Oxford DAVID SALESIN, Google Research WILLIAM T. FREEMAN, Google Research MICHAEL RUBINSTEIN, Google Research Input Video Frame t Frame t1 (child I jumps) Frame t2 (child II jumps) Frame t3 (child III jumps) Our Retiming Result Our Output Layers Frame t1 (all stand) Frame t2 (all jump) Frame t3 (all in water) Fig. 1. Making all children jump into the pool together — in post-processing! In the original video (left, top) each child is jumping into the poolata different time. In our computationally retimed video (left, bottom), the jumps of children I and III are time-aligned with that of child II, such thattheyalljump together into the pool (notice that child II remains unchanged in the input and output frames). In this paper, we present a method to produce this and other people retiming effects in natural, ordinary videos. Our method is based on a novel deep neural network that learns a layered decomposition oftheinput video (right). Our model not only disentangles the motions of people in different layers, but can also capture the various scene elements thatare correlated with those people (e.g., water splashes as the children hit the water, shadows, reflections). When people are retimed, those related elements get automatically retimed with them, which allows us to create realistic and faithful re-renderings of the video for a variety of retiming effects. The full input and retimed sequences are available in the supplementary video. -

ELEG 5173L Digital Signal Processing Ch. 3 Discrete-Time Fourier Transform
Department of Electrical Engineering University of Arkansas ELEG 5173L Digital Signal Processing Ch. 3 Discrete-Time Fourier Transform Dr. Jingxian Wu [email protected] 2 OUTLINE • The Discrete-Time Fourier Transform (DTFT) • Properties • DTFT of Sampled Signals • Upsampling and downsampling 3 DTFT • Discrete-time Fourier Transform (DTFT) X () x(n)e jn n – (radians): digital frequency • Review: Z-transform: X (z) x(n)zn n0 j X () X (z) | j – Replace z with e . ze • Review: Fourier transform: X () x(t)e jt – (rads/sec): analog frequency 4 DTFT • Relationship between DTFT and Fourier Transform – Sample a continuous time signal x a ( t ) with a sampling period T xs (t) xa (t) (t nT ) xa (nT ) (t nT ) n n – The Fourier Transform of ys (t) jt jnT X s () xs (t)e dt xa (nT)e n – Define: T • : digital frequency (unit: radians) • : analog frequency (unit: radians/sec) – Let x(n) xa (nT) X () X s T 5 DTFT • Relationship between DTFT and Fourier Transform (Cont’d) – The DTFT can be considered as the scaled version of the Fourier transform of the sampled continuous-time signal jt jnT X s () xs (t)e dt xa (nT)e n x(n) x (nT) T a jn X () X s x(n)e T n 6 DTFT • Discrete Frequency – Unit: radians (the unit of continuous frequency is radians/sec) – X ( ) is a periodic function with period 2 j2 n jn j2n jn X ( 2 ) x(n)e x(n)e e x(n)e X () n n n – We only need to consider for • For Fourier transform, we need to consider 1 – f T 2 2T 1 – f T 2 2T 7 DTFT • Example: find the DTFT of the following signal – 1. -
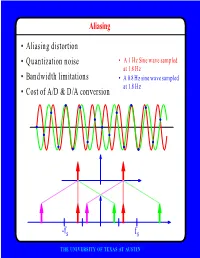
F • Aliasing Distortion • Quantization Noise • Bandwidth Limitations • Cost of A/D & D/A Conversion
Aliasing • Aliasing distortion • Quantization noise • A 1 Hz Sine wave sampled at 1.8 Hz • Bandwidth limitations • A 0.8 Hz sine wave sampled at 1.8 Hz • Cost of A/D & D/A conversion -fs fs THE UNIVERSITY OF TEXAS AT AUSTIN Advantages of Digital Systems Perfect reconstruction of a Better trade-off between signal is possible even after bandwidth and noise severe distortion immunity performance digital analog bandwidth Increase signal-to-noise ratio simply by adding more bits SNR = -7.2 + 6 dB/bit THE UNIVERSITY OF TEXAS AT AUSTIN Advantages of Digital Systems Programmability • Modifiable in the field • Implement multiple standards • Better user interfaces • Tolerance for changes in specifications • Get better use of hardware for low-speed operations • Debugging • User programmability THE UNIVERSITY OF TEXAS AT AUSTIN Disadvantages of Digital Systems Programmability • Speed is too slow for some applications • High average power and peak power consumption RISC (2 Watts) vs. DSP (50 mW) DATA PROG MEMORY MEMORY HARVARD ARCHITECTURE • Aliasing from undersampling • Clipping from quantization Q[v] v v THE UNIVERSITY OF TEXAS AT AUSTIN Analog-to-Digital Conversion 1 --- T h(t) Q[.] xt() yt() ynT() yˆ()nT Anti-Aliasing Sampler Quantizer Filter xt() y(nT) t n y(t) ^y(nT) t n THE UNIVERSITY OF TEXAS AT AUSTIN Resampling Changing the Sampling Rate • Conversion between audio formats Compact 48.0 Digital Disc ---------- Audio Tape 44.1 KHz44.1 48 KHz • Speech compression Speech 1 Speech for on DAT --- Telephone 48 KHz 6 8 KHz • Video format conversion -

Image Deconvolution with Deep Image Prior: Final Report
Image deconvolution with deep image prior: Final Report Abstract tion kernel is given (Sezan & Tekalp, 1990), i.e. recovering Image deconvolution has always been a hard in- X with knowing K in Equation 1. This problem is ill-posed, verse problem because of its mathematically ill- because simply applying the inverse of the convolution op- −1 posed property (Chan & Wong, 1998). A recent eration on degraded image B with kernel K, i.e. B∗ K, −1 work (Ulyanov et al., 2018) proposed deep im- gives an inverted noise term E∗ K, which dominates the age prior (DIP) which uses neural net structures solution (Hansen et al., 2006). to represent image prior information. This work Blind deconvolution: In reality, we can hardly obtain the mainly focuses on the task of image deconvolu- detailed kernel information, in which case the deconvolu- tion, using DIP to express image prior informa- tion problem is formulated in a blind setting (Kundur & tion and ref ne the domain of its objectives. It Hatzinakos, 1996). More concisely, blind deconvolution is proposes new energy functions for kernel-known to recover X without knowing K. This task is much more and blind deconvolution respectively in terms of challenging than it is under non-blind settings, because the DIP, and uses hourglass ConvNet (Newell et al., observed information becomes less and the domains of the 2016) as the DIP structures to represent sharpness variables become larger (Chan & Wong, 1998). or other higher level priors of natural images, as well as the prior in degradation kernels. From the In image deconvolution, prior information on unknown results on 6 standard test images, we f rst prove images and kernels (in blind settings) can signif cantly im- that DIP with ConvNet structure is strongly ca- prove the deconvolved results. -

PROCEEDINGS of the ICA CONGRESS (Onl the ICA PROCEEDINGS OF
ine) - ISSN 2415-1599 ISSN ine) - PROCEEDINGS OF THE ICA CONGRESS (onl THE ICA PROCEEDINGS OF Page intentionaly left blank 22nd International Congress on Acoustics ICA 2016 PROCEEDINGS Editors: Federico Miyara Ernesto Accolti Vivian Pasch Nilda Vechiatti X Congreso Iberoamericano de Acústica XIV Congreso Argentino de Acústica XXVI Encontro da Sociedade Brasileira de Acústica 22nd International Congress on Acoustics ICA 2016 : Proceedings / Federico Miyara ... [et al.] ; compilado por Federico Miyara ; Ernesto Accolti. - 1a ed . - Gonnet : Asociación de Acústicos Argentinos, 2016. Libro digital, PDF Archivo Digital: descarga y online ISBN 978-987-24713-6-1 1. Acústica. 2. Acústica Arquitectónica. 3. Electroacústica. I. Miyara, Federico II. Miyara, Federico, comp. III. Accolti, Ernesto, comp. CDD 690.22 ISSN 2415-1599 ISBN 978-987-24713-6-1 © Asociación de Acústicos Argentinos Hecho el depósito que marca la ley 11.723 Disclaimer: The material, information, results, opinions, and/or views in this publication, as well as the claim for authorship and originality, are the sole responsibility of the respective author(s) of each paper, not the International Commission for Acoustics, the Federación Iberoamaricana de Acústica, the Asociación de Acústicos Argentinos or any of their employees, members, authorities, or editors. Except for the cases in which it is expressly stated, the papers have not been subject to peer review. The editors have attempted to accomplish a uniform presentation for all papers and the authors have been given the opportunity -

Controlling Neural Networks Via Energy Dissipation
Controlling Neural Networks via Energy Dissipation Michael Moeller Thomas Mollenhoff¨ Daniel Cremers University of Siegen TU Munich TU Munich [email protected] [email protected] [email protected] Abstract and computing its Radon transform, respectively. Mathe- matically, the above problems can be phrased as linear in- The last decade has shown a tremendous success in verse problems in which one tries to recover the desired solving various computer vision problems with the help of quantity uˆ from measurements f that arise from applying deep learning techniques. Lately, many works have demon- an application-dependent linear operator A to the unknown strated that learning-based approaches with suitable net- and contain additive noise ξ: work architectures even exhibit superior performance for the solution of (ill-posed) image reconstruction problems f = Auˆ + ξ. (1) such as deblurring, super-resolution, or medical image re- Unfortunately, most practically relevant inverse problems construction. The drawback of purely learning-based meth- are ill-posed, meaning that equation (1) either does not de- ods, however, is that they cannot provide provable guaran- termine uˆ uniquely even if ξ = 0, or tiny amounts of noise tees for the trained network to follow a given data formation ξ can alter the naive prediction of uˆ significantly. These process during inference. In this work we propose energy phenomena have been well-investigated from a mathemat- dissipating networks that iteratively compute a descent di- ical perspective with regularization methods being the tool rection with respect to a given cost function or energy at the to still obtain provably stable reconstructions. -

2007 Registration Document
2007 REGISTRATION DOCUMENT (www.renault.com) REGISTRATION DOCUMENT REGISTRATION 2007 Photos cre dits: cover: Thomas Von Salomon - p. 3 : R. Kalvar - p. 4, 8, 22, 30 : BLM Studio, S. de Bourgies S. BLM Studio, 30 : 22, 8, 4, Kalvar - p. R. 3 : Salomon - p. Von Thomas cover: dits: Photos cre 2007 REGISTRATION DOCUMENT INCLUDING THE MANAGEMENT REPORT APPROVED BY THE BOARD OF DIRECTORS ON FEBRUARY 12, 2008 This Registration Document is on line on the website www .renault.com (French and English versions) and on the AMF website www .amf- france.org (French version only). TABLE OF CONTENTS 0 1 05 RENAULT AND THE GROUP 5 RENAULT AND ITS SHAREHOLDERS 157 1.1 Presentation of Renault and the Group 6 5.1 General information 158 1.2 Risk factors 24 5.2 General information about Renault’s share capital 160 1.3 The Renault-Nissan Alliance 25 5.3 Market for Renault shares 163 5.4 Investor relations policy 167 02 MANAGEMENT REPORT 43 06 2.1 Earnings report 44 MIXED GENERAL MEETING 2.2 Research and development 62 OF APRIL 29, 2008: PRESENTATION 2.3 Risk management 66 OF THE RESOLUTIONS 171 The Board first of all proposes the adoption of eleven resolutions by the Ordinary General Meeting 172 Next, six resolutions are within the powers of 03 the Extraordinary General Meeting 174 SUSTAINABLE DEVELOPMENT 79 Finally, the Board proposes the adoption of two resolutions by the Ordinary General Meeting 176 3.1 Employee-relations performance 80 3.2 Environmental performance 94 3.3 Social performance 109 3.4 Table of objectives (employee relations, environmental -

Denoising and Regularization Via Exploiting the Structural Bias of Convolutional Generators
Denoising and Regularization via Exploiting the Structural Bias of Convolutional Generators Reinhard Heckel∗ and Mahdi Soltanolkotabiy ∗Dept. of Electrical and Computer Engineering, Technical University of Munich yDept. of Electrical and Computer Engineering, University of Southern California October 31, 2019 Abstract Convolutional Neural Networks (CNNs) have emerged as highly successful tools for image generation, recovery, and restoration. This success is often attributed to large amounts of training data. However, recent experimental findings challenge this view and instead suggest that a major contributing factor to this success is that convolutional networks impose strong prior assumptions about natural images. A surprising experiment that highlights this architec- tural bias towards natural images is that one can remove noise and corruptions from a natural image without using any training data, by simply fitting (via gradient descent) a randomly initialized, over-parameterized convolutional generator to the single corrupted image. While this over-parameterized network can fit the corrupted image perfectly, surprisingly after a few iterations of gradient descent one obtains the uncorrupted image. This intriguing phenomena enables state-of-the-art CNN-based denoising and regularization of linear inverse problems such as compressive sensing. In this paper we take a step towards demystifying this experimental phenomena by attributing this effect to particular architectural choices of convolutional net- works, namely convolutions with fixed interpolating filters. We then formally characterize the dynamics of fitting a two layer convolutional generator to a noisy signal and prove that early- stopped gradient descent denoises/regularizes. This results relies on showing that convolutional generators fit the structured part of an image significantly faster than the corrupted portion.