Patterson Zanardini
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Papua New Guinea's Emergent Longline Fishery
Papua New Guinea's emergent longline fishery Two Hawaii-based fishing vessels are currently participating in the longline fishery in Papua New Guinea (PNG), in the south west tropical Pacific. The two vessels, which fish in the Northwestern Hawaiian Islands for lobsters are on charter in PNG between lobster fishing seasons. PNG is one of the largest Pacific nations, straddling Southeast Asia and the South Pacific, with one of the largest EEZs in the region and abundant tuna resources. Like the rest of the Pacific, PNG is keen to expand its longline fishing industry for the lucrative fresh tuna market in Japan. Fishing trials in 1994 and 1995 in Rabaul, Finchaven and the capital city, Port Moresby, demonstrated the feasibility of a domestic fishery in PNG; although the productivity of the resource was evident from the long history of fishing in PNG waters by Korean, Taiwanese and Japanese longliners. From 1994 onwards various local companies began to establish fishing operations at various ports throughout the country, but with most activity centered around Port Moresby. A longline fishery management plan was developed by the National Fisheries Authority in 1995, which included a ban on foreign longliners operating in the PNG EEZ. Licensing guidelines were also introduced which included a minimum of 51% PNG equity in joint ventures, and short-term charter of foreign vessels to PNG companies on a 1:1 basis with the number of local vessels in a company fleet. Presently there are 20 longliners operating in PNG, with the majority of vessels being based in Port Moresby. Fishing companies outside of the capital include one in Alotau and the two others in PNG=s second city Lae. -

Papua New Guinea
PAPUA NEW GUINEA EMERGENCY PREPAREDNESS OPERATIONAL LOGISTICS CONTINGENCY PLAN PART 2 –EXISTING RESPONSE CAPACITY & OVERVIEW OF LOGISTICS SITUATION GLOBAL LOGISTICS CLUSTER – WFP FEBRUARY – MARCH 2011 1 | P a g e A. Summary A. SUMMARY 2 B. EXISTING RESPONSE CAPACITIES 4 C. LOGISTICS ACTORS 6 A. THE LOGISTICS COORDINATION GROUP 6 B. PAPUA NEW GUINEAN ACTORS 6 AT NATIONAL LEVEL 6 AT PROVINCIAL LEVEL 9 C. INTERNATIONAL COORDINATION BODIES 10 DMT 10 THE INTERNATIONAL DEVELOPMENT COUNCIL 10 D. OVERVIEW OF LOGISTICS INFRASTRUCTURE, SERVICES & STOCKS 11 A. LOGISTICS INFRASTRUCTURES OF PNG 11 PORTS 11 AIRPORTS 14 ROADS 15 WATERWAYS 17 STORAGE 18 MILLING CAPACITIES 19 B. LOGISTICS SERVICES OF PNG 20 GENERAL CONSIDERATIONS 20 FUEL SUPPLY 20 TRANSPORTERS 21 HEAVY HANDLING AND POWER EQUIPMENT 21 POWER SUPPLY 21 TELECOMS 22 LOCAL SUPPLIES MARKETS 22 C. CUSTOMS CLEARANCE 23 IMPORT CLEARANCE PROCEDURES 23 TAX EXEMPTION PROCESS 24 THE IMPORTING PROCESS FOR EXEMPTIONS 25 D. REGULATORY DEPARTMENTS 26 CASA 26 DEPARTMENT OF HEALTH 26 NATIONAL INFORMATION AND COMMUNICATIONS TECHNOLOGY AUTHORITY (NICTA) 27 2 | P a g e MARITIME AUTHORITIES 28 1. NATIONAL MARITIME SAFETY AUTHORITY 28 2. TECHNICAL DEPARTMENTS DEPENDING FROM THE NATIONAL PORT CORPORATION LTD 30 E. PNG GLOBAL LOGISTICS CONCEPT OF OPERATIONS 34 A. CHALLENGES AND SOLUTIONS PROPOSED 34 MAJOR PROBLEMS/BOTTLENECKS IDENTIFIED: 34 SOLUTIONS PROPOSED 34 B. EXISTING OPERATIONAL CORRIDORS IN PNG 35 MAIN ENTRY POINTS: 35 SECONDARY ENTRY POINTS: 35 EXISTING CORRIDORS: 36 LOGISTICS HUBS: 39 C. STORAGE: 41 CURRENT SITUATION: 41 PROPOSED LONG TERM SOLUTION 41 DURING EMERGENCIES 41 D. DELIVERIES: 41 3 | P a g e B. Existing response capacities Here under is an updated list of the main response capacities currently present in the country. -

PCRAFI AIR Brochure- Papua New Guinea (1).Pdf
PACIFIC CATASTROPHE RISK ASSESSMENT AND FINANCING INITIATIVE Public Disclosure Authorized PAPUA NEW GUINEA Public Disclosure Authorized SEPTEMBER 2011 COUNTRY RISK PROFILE: PAPUA NEW GUINEA Papua New Guinea is expected to incur, on average, 85 million USD per year in losses due to earthquakes and Public Disclosure Authorized tropical cyclones. In the next 50 years, Papua New Guinea has a 50% chance of experiencing a loss exceeding 700 million USD and casualties larger than 4,900 people, and a 10% chance of experiencing a loss exceeding 1.4 billion USD and casualties larger than 11,500 people. Public Disclosure Authorized BETTER RISK INFORMATION FOR SMARTER INVESTMENTS COUNTRY RISK PROFILE: PAPAU NEW GUINEA POPULATION, BUILDINGS, INFRASTRUCTURE AND CROPS EXPOSED TO NATURAL PERILS An extensive study has been conducted to assemble a comprehensive inventory of population and properties at risk. Properties include residential, commercial, public and industrial buildings; infrastructure assets such as major ports, airports, power plants, bridges, and roads; and major crops, such as coconut, palm oil, taro, sugar cane and many others. TABLE 1: Summary of Exposure in Papau New Guinea (2010) General Information: Total Population: 6,406,000 GDP Per Capita (USD): 1,480 Total GDP (million USD): 9,480.0 Asset Counts: Figure 1: Building locations. Residential Buildings: 2,261,485 145° E 150° E 155° E Public Buildings: 43,258 0 100 200 400 Commercial, Industrial, and Other Buildings: 88,536 Kilometers All Buildings: 2,393,279 Hectares of Major Crops: 1,350,990 S S ° ° Cost of Replacing Assets (million USD): 5 5 Buildings: 39,509 Infrastructure: 6,639 Lae Crops: 3,061 Port Moresby Total: 49,209 S S ° ° 0 0 Government Revenue and Expenditure: 1 Building Replacement Cost 1 Density (million USD / km^2) Total Government Revenue 0 - 0.025 0.075 - 0.1 0.5 - 1 0.025 - 0.05 0.1 - 0.25 1 - 156 (Million USD): 2,217.9 0.05 - 0.075 0.25 - 0.5 Papua New Guinea 145° E 150° E 155° E (% GDP): 23.4% Figure 2: Building replacement cost density by district. -

Papua New Guinea Trans-Island Highway Construction Project (I) (II) Report Date: February 2003 Field Survey: October – November 2002 1
Papua New Guinea Trans-Island Highway Construction Project (I) (II) Report Date: February 2003 Field Survey: October – November 2002 1. Project Profile and Japan’s ODA Loan Wewack Indonesia Papua New Guinea Lae Project site Port Moresby Australia Project site Section of the Bereina-Malalaua Road 1.1 Background In Papua New Guinea (PNG), the absence of road links between its capital, Port Moresby, the second city, Lae, and the Highland Province, home to vigorous mining and agricultural production activity, meant a sole reliance on marine and air transport, a situation that was highly inefficient both in economic and social terms. To address this, the government formulated plans to construct a road linking the cities of Port Moresby and Lae that would pass through the Owen Stanley mountains, and in 1979, the Australian government supported the implementation of a feasibility study (F/S) for this route. The plan for the trans-island Port Moresby – Lae highway comprised a stretch spanning 575 kilometers, and included construction of a new 80 kilometer road between Bereina and Malalaua, detailed designs for the 135 kilometer section between Malalaua and Aseki, and widening of the existing 90 kilometer stretch between Aseki and Latep (all distances are approximate). Other sections were excluded from the plan. In the southern Bereina-Malalaua section that was covered by this project, local residents were dependent upon marine transport, a situation that was, in fact, akin to living on an isolated inland islands. It was anticipated that the construction of a new road linking the area with the capital Port Moresby, would make a substantial contribution to economic development and to improving the services like health and education. -

47356-002: Power Sector Development Investment Program
Initial Environmental Examination March 2021 Papua New Guinea: Power Sector Development Project Prepared by PNG Power Ltd for the Department of Petroleum and Energy, PNG, and the Asian Development Bank CURRENCY EQUIVALENTS (as of 10 March 2021) Currency unit – kina (K) K1.00 = $0.29 $1.00 = K3.51 ABBREVIATIONS ADB – Asian Development Bank AIFFP – Australian Infrastructure Financing Facility for the Pacific CEMP – Contractor Environmental Management Plan DNPM – Department of National Planning and Monitoring EMP – environment management plan FIRR – financial internal rate of return GBV – gender-based violence IEE – initial environmental examination kV – kilovolt km – kilometer MW – megawatt PAM – project administration manual PMU – project management unit PNG – Papua New Guinea PPL – PNG Power Limited ROW – right-of-way SCADA – supervisory control and data acquisition SOE – state-owned enterprise STEM – Science, Technology, Engineering, and Mathematics TEIP – Town Electrification Investment Program NOTE In this report, “$” refers to United States dollars unless otherwise stated. This initial environmental examination is a document of the borrower. The views expressed herein do not necessarily represent those of ADB's Board of Directors, Management, or staff, and may be preliminary in nature. Your attention is directed to the “terms of use” section of this website. In preparing any country program or strategy, financing any project, or by making any designation of or reference to a particular territory or geographic area in this document, the Asian ii Development Bank does not intend to make any judgments as to the legal or other status of any territory or area. iii CONTENTS CONTENTS I FIGURES II TABLES II ABBREVIATIONS III ELECTRICAL TERMINOLOGY III EXECUTIVE SUMMARY V I. -

Harvesting Development
HARVESTING DEVELOPMENT The Nordic Institute of Asian Studies (NIAS) is funded by the govern- ments of Denmark, Finland, Iceland, Norway and Sweden via the Nordic Council of Ministers, and works to encourage and support Asian studies in the Nordic countries. In so doing, NIAS has been publishing books since 1969, with more than one hundred titles produced in the last decade. Nordic Council of Ministers HARVESTING DEVELOPMENT THE CONSTRUCTION OF FRESH FOOD MARKETS IN PAPUA NEW GUINEA Karl Benediktsson Copyright © Karl Benediktsson 2002 All rights reserved. First Published in Denmark by Nordic Institute of Asian Studies (Simultaneously published in North America by The University of Michigan Press) Printed in Singapore No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, or otherwise, without the written permission of the publisher. British Library Cataloguing in Publication Data Benediktsson, Karl Harvesting development : the construction of fresh food markets in Papua New Guinea 1.Food supply - Papua New Guinea 2.Farm produce - Papua New Guinea I.Title II.Nordic Institute of Asian Studies 381'.4'5'6413'009953 ISBN 87-87062-92-5 (cloth) ISBN 87-87062-91-7 (paper) Contents Illustrations … vi Tables … viii Vignettes … viii Acknowledgements … ix Abbreviations … xii 1Introduction … 1 2Markets, commoditization, and actors: spacious concepts … 22 3Faces in the crowd: Lives and networks of selected actors … 54 4Fresh food movements in a fragmented national -

Wilson Kamit: Current Economic and Financial Conditions in Papua New
Wilson Kamit: Current economic and financial conditions in Papua New Guinea Speech by Mr Wilson Kamit, Governor of the Bank of Papua New Guinea, to the Western Highlands Provincial Community, on the occasion of the Bank Board meeting, Mt Hagen, 15 June 2005. * * * 1. Introduction Representatives of the Western Highlands Provincial Administration, leaders of the business and civic sectors of the Western Highlands community, distinguished guests, ladies and gentlemen. On behalf of the Board and officers of the Bank of Papua New Guinea, I wish to express our great pleasure for us to be here and to have the opportunity to meet with you. Many of you are no strangers to the Central Bank through the Business Liaison Surveys you assist in. The members of the Board comprise the Governor as Chairman, the two Deputy Governors, Mr. Benny Popoitai, the Deputy Governor for Management and Operations, and Mr. Loi Bakani, the Deputy Governor for Policy and Regulations. The members of the Board (ex-officio) who are appointed in accordance with Section 27 of the CBA (2000), which specifies representation by various industry and community organisations are: Mr. John Mahuk, President of the PNG Trade Union Congress Mr. Michael Mayberry, President of the PNG Chamber of Commerce Mr. Kostas Constantinou – Managing Director of the Lamana Hotel Ms. Betty Palaso – Deputy Commissioner, IRC There are three vacancies, the President of the PNG Institute of Accountants, the Chairman of the Securities Commission and another appointee of the Minister. The vacancy in the PNG Institute of Accountants was due to the death of the son of the Western Highlands Province, the Late Mr Patrick Kolta. -

Annual Evaluation July 01, 1984
ANNUAL EVALUATION JULY 01, 1984 - SEPTEMBER 30, 1985 USAID OPG 879-0001-G-40099 A PROGRAM OF INSTITUTION BUILDING FOR SMALL BUSINESS DEVELOPMENT THE FOUNDATION FOR THE PEOPLES OF THE SOUTH PACIFIC PACIFIC HOUSE POBOX 727 2-12 WEST PARK AVENUE LONG BEACH, N,Y,11561 INDEX INTRODUCTION: Page I ADMINISTRATION 1 II SPATF 2 (a) Introduction 2 (b) SPATF HQ 3 (c) SPATF Handicrafts 3 (d) VES 4 (e) ATDI 4 (f) Hohola Small Industries and Hanuatek 6 III SMALL GRANTS PROGRAM FOR COTTAGE INDUSTRIES 7 IV HOHCLA YOUTH DEVELOPMENT CENTER 10 V EVALUATION 12 VI THE FUTURE 13 VII RECOMMENDATIONS 14 APPENDIX I INTRODUCTION The present Operational Program Grant was signed on July 31, 1984, effective from July 1, 1984. It is an entirely new program; virtually all the previous programs were phased out. This Evaluation was carried out by Dr.Bernard P.Hosie, the Chief Evaluator for the Foundation for the Peoples of the South Pacific (FSP) between October 6 and October 17. It covers the period July 1, 1984 to September 30, 1985. The evaluator concentrated on Madang, Lae and Port Moresby, with only one side trip to a project in the Highlands. Madang is the office of the FSP Country Director. FSP has an office in the Appropriate Technology Development Institute (ATDI) in Lae, staffed by the Assistant Country Director and four Extension Officers. Port Moresby is Headquarters for the South Pacific Appropriate Technology Foundation (SPATF) the main Papua New Guinea partner of FSP. i ADMINISTRATION The FSP Headquarters in Papun New Guinea is in Madang, in two rooms rented from the Lutheran Kristen Press. -

Family Protection Orders in Papua New Guinea: Summary Report Judy Putt and Lindy Kanan
Family Protection Orders in Papua New Guinea: Summary Report Judy Putt and Lindy Kanan Introduction The findings from the research are synthesised and Funded by the Australian Government through the summarised under the headings implementation and Pacific Women Shaping Pacific Development program, impact. Justice Services and Stability for Development program Implementation and the Pacific Research Program, the purpose of our The stated intent of the FPA includes promoting safe, research project was to investigate whether the family stable and strong families; preventing and deterring protection orders (FPOs) introduced under the Family domestic violence at all levels of society; and ensuring Protection Act 2013 (FPA) were being accessed by, and there is effective protection for the victims of domestic improving the safety of, domestic and family violence1 violence. Upon application, an FPO can be issued by (DFV) survivors. The Act, as well as the introduction of a court for a stipulated period of time, with conditions civil restraining orders specifically targeted at preventing imposed on the respondent that seek to prevent further and reducing DFV, represented a significant milestone in domestic violence. Those who can apply for FPOs efforts to address in DFV in Papua New Guinea (PNG). include spouses, family members — including children The research project on FPOs in PNG was undertaken — and any person treated by the spouse as a family from late 2019 to August 2020.2 It was a collaborative member. Regulations for the FPA were gazetted, and project that involved researchers based in Australia and guidelines produced by the Department of Justice and PNG, local researchers in various communities and the Attorney General, in 2017. -

NATIONAL FISHERIES AUTHORITY Office of Tlie 9,1.Anaging <Director
NATIONALFISHERIES AUTHORITY Officeof tlie 9,1.anaging <Director Telephone: (675) 309 0444 Level 11, Kina Bank Haus Facsimile: (675) 3202061 Douglas Street Email: [email protected] PO Box 2016 Website: www.fisheries.gov.pg Port Moresby, N.C.D Papua New Guinea CIRCULAR No. 09/2020 TO ALL FISH PROCESSING FACILITY OPERATORS, LICENSED VESSELS FISHING FOR 2020 LICENSING PERIOD. FROM John E. Kasu, Managing Director DATE 23rd March 2020 SUBJECT : MANDATORY MEASURES TO MITIGATE THE SPREAD OF THE NOVEL CORONAVIRUS (COVID-19) The Government has declared a State of Emergency starting 24th March 2020 per the Official Statement from the Prime Minister's Office dated 22nd March 2020. The Government of Papua New Guinea has heightened surveillance since the first case of COVID-19. This is Circular No.3 in an effort to mitigate the spread of this deadly virus. This circular supersedes the prev10us circulars sent out by the National Fisheries Authority. All Fishing Companies, Fishing Vessel Operators and Fish Processing Facility Operators are directed to; 1) Adhere to the measures outlined in the Prime Ministers Official Statement in declaring a State of Emergency. 2) Adhere to all measures and requirements set by the Department of Health, Ministry of Immigration and Border Security, Department of Transport and other Departments and Agencies of the PNG Government. 3) All foreign crew and employees who have flown into PNG after 6th March 2020 are to be identified and isolated for testing by the National National Fisheries College Lae Office Maclang Office -
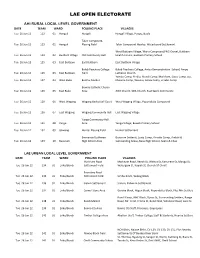
Lae Open Electorate
LAE OPEN ELECTORATE AHI RURAL LOCAL LEVEL GOVERNMENT DATE TEAM WARD POLLING PLACE VILLAGES Tue 26 Jun 12 122 01 Hengali Hengali Hengali Village, Poapu, Buala Talair Compound, Tue 26 Jun 12 123 01 Hengali Playing Field Talair Compound Nearby, Waterboard Settlement West Buitbam Village, Waria Compound,PNG Gravel, Buitbam Tue 26 Jun 12 124 02 Buitbam Village Old Community Hall Health Centre, Buitbam Primary School Tue 26 Jun 12 125 03 East Buitbam East Buitbam East Buitbam Village Balob Teachers College Balob Teachers College, Amba Demonstration School, Ampo Tue 26 Jun 12 126 03 East Buitbam Field Lutheran Church Yambo Comp, Pindiu, Mendi Comp, Markham, Siassi Comp, Sio, Tue 26 Jun 12 127 04 West Buko Bumbu Market Maiama Comp, Woseta, Amoa Comp, Zinabe Comp Bumbu Catholic Church Tue 26 Jun 12 128 05 East Buko Area AOG Church, SDA Church, East Sepik Community Tue 26 Jun 12 129 06 West Wagang Wagang Basketball Court West Wagang Village, Popondetta Compound Tue 26 Jun 12 130 07 East Wagang Wagang Community Hall East Wagang Village Yanga Community Hall Tue 26 Jun 12 131 08 Yanga Area Yanga Village, Bowali Primary School Tue 26 Jun 12 132 09 Gawang Hunter Playing Field Hunter Settlement Emmanuel Lutheran Busurum Settmnt, Lusip Comp, Arnotts Comp, Ambisi & Tue 26 Jun 12 133 10 Busurum High School Area Surrounding Areas, Busu High School, Seeto & Chan LAE URBAN LOCAL LEVEL GOVERNMENT DATE TEAM WARD POLLING PLACE VILLAGES Markham Road Markham Road, Beech St, Walnut St, Kamarere St, Mango St, Tue 26 Jun 12 134 01 Eriku/Bundi Settlement Field Watergum St, Kapiak St, Church Of Christ Boundary Road Tue 26 Jun 12 135 01 Eriku/Bundi Settlement Field Simbu Block, Wabag Block Tue 26 Jun 12 136 01 Eriku/Bundi Sialum Settlement Sialum, Kabwum Settlement Tue 26 Jun 12 137 01 Eriku/Bundi Corner Store Area Goroka Block, Hagen Block, Popondetta Block, Plus Mix Settlers Bundi Comp, NHC Block, Dysox St, Surrounding Settlers, Range Tue 26 Jun 12 138 01 Eriku/Bundi Bundi Market Road, Mr. -

Kuantim Mi Tu (“Count Me Too”): Using Multiple Methods to Estimate The
JMIR PUBLIC HEALTH AND SURVEILLANCE Weikum et al Original Paper Kuantim mi tu (ªCount me tooº): Using Multiple Methods to Estimate the Number of Female Sex Workers, Men Who Have Sex With Men, and Transgender Women in Papua New Guinea in 2016 and 2017 Damian Weikum1, MPH; Angela Kelly-Hanku2,3, PhD; Parker Hou3; Martha Kupul3, BS; Angelyne Amos-Kuma3, MIPH; Steven G Badman2, MPH, JP; Nick Dala4, MBBS, MMed; Kelsey C Coy1, MPH; John M Kaldor2, PhD; Andrew J Vallely2, MBBS, MRCP, DTM&H, MSc, DLSHTM, PhD; Avi J Hakim1, MA, MPH 1US Centers for Disease Control and Prevention, Atlanta, GA, United States 2Kirby Institute, UNSW, Sydney, Australia 3Papua New Guinea Institute of Medical Research, Goroka, Papua New Guinea 4Papua New Guinea National Department of Health, Port Moresby, Papua New Guinea Corresponding Author: Damian Weikum, MPH US Centers for Disease Control and Prevention 1600 Clifton Road, NE Mail Stop E-30 Atlanta, GA, 30329 United States Phone: 1 4047185688 Email: [email protected] Abstract Background: Female sex workers (FSW), men who have sex with men (MSM), and transgender women (TGW) are at high risk of acquiring HIV in many settings, such as Papua New Guinea (PNG). An understanding of the approximate size of these populations can inform resource allocation for HIV services for FSW, MSM, and TGW. Objective: An objective of this multi-site survey was to conduct updated population size estimations (PSE) of FSW and MSM/TGW. Methods: Respondent-driven sampling (RDS) biobehavioral surveys of FSW and MSM/TGW were conducted in 3 major citiesÐ(1) Port Moresby, (2) Lae, and (3) Mount HagenÐbetween June 2016 and December 2017.