A Whole-Brain Connectivity Map of Mouse Insular Cortex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Five Topographically Organized Fields in the Somatosensory Cortex of the Flying Fox: Microelectrode Maps, Myeloarchitecture, and Cortical Modules
THE JOURNAL OF COMPARATIVE NEUROLOGY 317:1-30 (1992) Five Topographically Organized Fields in the Somatosensory Cortex of the Flying Fox: Microelectrode Maps, Myeloarchitecture, and Cortical Modules LEAH A. KRUBITZER AND MIKE B. CALFORD Vision, Touch and Hearing Research Centre, Department of Physiology and Pharmacology, The University of Queensland, Queensland, Australia 4072 ABSTRACT Five somatosensory fields were defined in the grey-headed flying fox by using microelec- trode mapping procedures. These fields are: the primary somatosensory area, SI or area 3b; a field caudal to area 3b, area 1/2; the second somatosensory area, SII; the parietal ventral area, PV; and the ventral somatosensory area, VS. A large number of closely spaced electrode penetrations recording multiunit activity revealed that each of these fields had a complete somatotopic representation. Microelectrode maps of somatosensory fields were related to architecture in cortex that had been flattened, cut parallel to the cortical surface, and stained for myelin. Receptive field size and some neural properties of individual fields were directly compared. Area 3b was the largest field identified and its topography was similar to that described in many other mammals. Neurons in 3b were highly responsive to cutaneous stimulation of peripheral body parts and had relatively small receptive fields. The myeloarchi- tecture revealed patches of dense myelination surrounded by thin zones of lightly myelinated cortex. Microelectrode recordings showed that myelin-dense and sparse zones in 3b were related to neurons that responded consistently or habituated to repetitive stimulation respectively. In cortex caudal to 3b, and protruding into 3b, a complete representation of the body surface adjacent to much of the caudal boundary of 3b was defined. -

Anatomy of the Temporal Lobe
Hindawi Publishing Corporation Epilepsy Research and Treatment Volume 2012, Article ID 176157, 12 pages doi:10.1155/2012/176157 Review Article AnatomyoftheTemporalLobe J. A. Kiernan Department of Anatomy and Cell Biology, The University of Western Ontario, London, ON, Canada N6A 5C1 Correspondence should be addressed to J. A. Kiernan, [email protected] Received 6 October 2011; Accepted 3 December 2011 Academic Editor: Seyed M. Mirsattari Copyright © 2012 J. A. Kiernan. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Only primates have temporal lobes, which are largest in man, accommodating 17% of the cerebral cortex and including areas with auditory, olfactory, vestibular, visual and linguistic functions. The hippocampal formation, on the medial side of the lobe, includes the parahippocampal gyrus, subiculum, hippocampus, dentate gyrus, and associated white matter, notably the fimbria, whose fibres continue into the fornix. The hippocampus is an inrolled gyrus that bulges into the temporal horn of the lateral ventricle. Association fibres connect all parts of the cerebral cortex with the parahippocampal gyrus and subiculum, which in turn project to the dentate gyrus. The largest efferent projection of the subiculum and hippocampus is through the fornix to the hypothalamus. The choroid fissure, alongside the fimbria, separates the temporal lobe from the optic tract, hypothalamus and midbrain. The amygdala comprises several nuclei on the medial aspect of the temporal lobe, mostly anterior the hippocampus and indenting the tip of the temporal horn. The amygdala receives input from the olfactory bulb and from association cortex for other modalities of sensation. -

Interoception: the Sense of the Physiological Condition of the Body AD (Bud) Craig
500 Interoception: the sense of the physiological condition of the body AD (Bud) Craig Converging evidence indicates that primates have a distinct the body share. Recent findings that compel a conceptual cortical image of homeostatic afferent activity that reflects all shift resolve these issues by showing that all feelings from aspects of the physiological condition of all tissues of the body. the body are represented in a phylogenetically new system This interoceptive system, associated with autonomic motor in primates. This system has evolved from the afferent control, is distinct from the exteroceptive system (cutaneous limb of the evolutionarily ancient, hierarchical homeostatic mechanoreception and proprioception) that guides somatic system that maintains the integrity of the body. These motor activity. The primary interoceptive representation in the feelings represent a sense of the physiological condition of dorsal posterior insula engenders distinct highly resolved the entire body, redefining the category ‘interoception’. feelings from the body that include pain, temperature, itch, The present article summarizes this new view; more sensual touch, muscular and visceral sensations, vasomotor detailed reviews are available elsewhere [1,2]. activity, hunger, thirst, and ‘air hunger’. In humans, a meta- representation of the primary interoceptive activity is A homeostatic afferent pathway engendered in the right anterior insula, which seems to provide Anatomical characteristics the basis for the subjective image of the material self as a feeling Cannon [3] recognized that the neural processes (auto- (sentient) entity, that is, emotional awareness. nomic, neuroendocrine and behavioral) that maintain opti- mal physiological balance in the body, or homeostasis, must Addresses receive afferent inputs that report the condition of the Atkinson Pain Research Laboratory, Division of Neurosurgery, tissues of the body. -

The Contribution of Sensory System Functional Connectivity Reduction to Clinical Pain in Fibromyalgia
1 The contribution of sensory system functional connectivity reduction to clinical pain in fibromyalgia Jesus Pujol1,2, Dídac Macià1, Alba Garcia-Fontanals3, Laura Blanco-Hinojo1,4, Marina López-Solà1,5, Susana Garcia-Blanco6, Violant Poca-Dias6, Ben J Harrison7, Oren Contreras-Rodríguez1, Jordi Monfort8, Ferran Garcia-Fructuoso6, Joan Deus1,3 1MRI Research Unit, CRC Mar, Hospital del Mar, Barcelona, Spain. 2Centro Investigación Biomédica en Red de Salud Mental, CIBERSAM G21, Barcelona, Spain. 3Department of Clinical and Health Psychology, Autonomous University of Barcelona, Spain. 4Human Pharmacology and Neurosciences, Institute of Neuropsychiatry and Addiction, Hospital del Mar Research Institute, Barcelona, Spain. 5Department of Psychology and Neuroscience. University of Colorado, Boulder, Colorado. 6Rheumatology Department, Hospital CIMA Sanitas, Barcelona, Spain. 7Melbourne Neuropsychiatry Centre, Department of Psychiatry, The University of Melbourne, Melbourne, Australia. 8Rheumatology Department, Hospital del Mar, Barcelona, Spain The submission contains: The main text file (double-spaced 39 pages) Six Figures One supplemental information file Corresponding author: Dr. Jesus Pujol Department of Magnetic Resonance, CRC-Mar, Hospital del Mar Passeig Marítim 25-29. 08003, Barcelona, Spain Email: [email protected] Telephone: + 34 93 221 21 80 Fax: + 34 93 221 21 81 Synopsis/ Summary Clinical pain in fibromyalgia is associated with functional changes at different brain levels in a pattern suggesting a general weakening of sensory integration. 2 Abstract Fibromyalgia typically presents with spontaneous body pain with no apparent cause and is considered pathophysiologically to be a functional disorder of somatosensory processing. We have investigated potential associations between the degree of self-reported clinical pain and resting-state brain functional connectivity at different levels of putative somatosensory integration. -
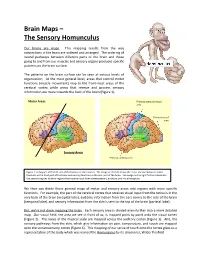
Brain Maps – the Sensory Homunculus
Brain Maps – The Sensory Homunculus Our brains are maps. This mapping results from the way connections in the brain are ordered and arranged. The ordering of neural pathways between different parts of the brain and those going to and from our muscles and sensory organs produces specific patterns on the brain surface. The patterns on the brain surface can be seen at various levels of organization. At the most general level, areas that control motor functions (muscle movement) map to the front-most areas of the cerebral cortex while areas that receive and process sensory information are more towards the back of the brain (Figure 1). Motor Areas Primary somatosensory area Primary visual area Sensory Areas Primary auditory area Figure 1. A diagram of the left side of the human cerebral cortex. The image on the left shows the major division between motor functions in the front part of the brain and sensory functions in the rear part of the brain. The image on the right further subdivides the sensory regions to show regions that receive input from somatosensory, auditory, and visual receptors. We then can divide these general maps of motor and sensory areas into regions with more specific functions. For example, the part of the cerebral cortex that receives visual input from the retina is in the very back of the brain (occipital lobe), auditory information from the ears comes to the side of the brain (temporal lobe), and sensory information from the skin is sent to the top of the brain (parietal lobe). But, we’re not done mapping the brain. -

Revista Brasileira De Psiquiatria Official Journal of the Brazilian Psychiatric Association Psychiatry Volume 34 • Number 1 • March/2012
Rev Bras Psiquiatr. 2012;34:101-111 Revista Brasileira de Psiquiatria Official Journal of the Brazilian Psychiatric Association Psychiatry Volume 34 • Number 1 • March/2012 REVIEW ARTICLE Neuroimaging in specific phobia disorder: a systematic review of the literature Ila M.P. Linares,1 Clarissa Trzesniak,1 Marcos Hortes N. Chagas,1 Jaime E. C. Hallak,1 Antonio E. Nardi,2 José Alexandre S. Crippa1 ¹ Department of Neuroscience and Behavior of the Ribeirão Preto Medical School, Universidade de São Paulo (FMRP-USP). INCT Translational Medicine (CNPq). São Paulo, Brazil 2 Panic & Respiration Laboratory. Institute of Psychiatry, Universidade Federal do Rio de Janeiro (UFRJ). INCT Translational Medicine (CNPq). Rio de Janeiro, Brazil Received on August 03, 2011; accepted on October 12, 2011 DESCRIPTORS Abstract Neuroimaging; Objective: Specific phobia (SP) is characterized by irrational fear associated with avoidance of Specific Phobia; specific stimuli. In recent years, neuroimaging techniques have been used in an attempt to better Review; understand the neurobiology of anxiety disorders. The objective of this study was to perform a Anxiety Disorder; systematic review of articles that used neuroimaging techniques to study SP. Method: A literature Phobia. search was conducted through electronic databases, using the keywords: imaging, neuroimaging, PET, spectroscopy, functional magnetic resonance, structural magnetic resonance, SPECT, MRI, DTI, and tractography, combined with simple phobia and specific phobia. One-hundred fifteen articles were found, of which 38 were selected for the present review. From these, 24 used fMRI, 11 used PET, 1 used SPECT, 2 used structural MRI, and none used spectroscopy. Result: The search showed that studies in this area were published recently and that the neuroanatomic substrate of SP has not yet been consolidated. -

Function of Cerebral Cortex
FUNCTION OF CEREBRAL CORTEX Course: Neuropsychology CC-6 (M.A PSYCHOLOGY SEM II); Unit I By Dr. Priyanka Kumari Assistant Professor Institute of Psychological Research and Service Patna University Contact No.7654991023; E-mail- [email protected] The cerebral cortex—the thin outer covering of the brain-is the part of the brain responsible for our ability to reason, plan, remember, and imagine. Cerebral Cortex accounts for our impressive capacity to process and transform information. The cerebral cortex is only about one-eighth of an inch thick, but it contains billions of neurons, each connected to thousands of others. The predominance of cell bodies gives the cortex a brownish gray colour. Because of its appearance, the cortex is often referred to as gray matter. Beneath the cortex are myelin-sheathed axons connecting the neurons of the cortex with those of other parts of the brain. The large concentrations of myelin make this tissue look whitish and opaque, and hence it is often referred to as white matter. The cortex is divided into two nearly symmetrical halves, the cerebral hemispheres . Thus, many of the structures of the cerebral cortex appear in both the left and right cerebral hemispheres. The two hemispheres appear to be somewhat specialized in the functions they perform. The cerebral hemispheres are folded into many ridges and grooves, which greatly increase their surface area. Each hemisphere is usually described, on the basis of the largest of these grooves or fissures, as being divided into four distinct regions or lobes. The four lobes are: • Frontal, • Parietal, • Occipital, and • Temporal. -

Microsurgical and Tractographic Anatomical Study of Insular and Transsylvian Transinsular Approach
Neurol Sci (2011) 32:865–874 DOI 10.1007/s10072-011-0721-2 ORIGINAL ARTICLE Microsurgical and tractographic anatomical study of insular and transsylvian transinsular approach Feng Wang • Tao Sun • XinGang Li • HeChun Xia • ZongZheng Li Received: 29 September 2008 / Accepted: 16 July 2011 / Published online: 24 August 2011 Ó The Author(s) 2011. This article is published with open access at Springerlink.com Abstract This study is to define the operative anatomy of Introduction the insula with emphasis on the transsylvian transinsular approach. The anatomy was studied in 15 brain specimens, In humans, the insula is a highly developed structure, among five were dissected by use of fiber dissection totally encased within the brain. In many clinical and technique; diffusion tensor imaging of 10 healthy volun- experimental studies, a variety of functions have been teers was obtained with a 1.5-T MR system. The temporal attributed to the insula, however, the full and comprehen- stem consists mainly of the uncinate fasciculus, inferior sive role that it plays continues to remain obscure. Oper- occipitofrontal fasciculus, Meyer’s loop of the optic radi- ation of neurosurgery, specifically of epilepsy surgery, is a ation and anterior commissure. The transinsular approach window onto function and dysfunction of the human brain requires an incision of the inferior limiting sulcus. In this [1]. The insula, as part of the paralimbic system, has both procedure, the fibers of the temporal stem can be inter- invasive anatomical and functional connections with the rupted to various degrees. The fiber dissection technique is temporal lobe through white matter fibers [2–6]. -

Altered Resting State Connectivity of the Insular Cortex in Individuals with Fibromyalgia
The Journal of Pain, Vol 15, No 8 (August), 2014: pp 815-826 Available online at www.jpain.org and www.sciencedirect.com Original Reports Altered Resting State Connectivity of the Insular Cortex in Individuals With Fibromyalgia Eric Ichesco,* Tobias Schmidt-Wilcke,*,** Rupal Bhavsar,*,y Daniel J. Clauw,* Scott J. Peltier,z Jieun Kim,x Vitaly Napadow,x,{,k Johnson P. Hampson,* Anson E. Kairys,*,yy David A. Williams,* and Richard E. Harris* *Department of Anesthesiology, Chronic Pain and Fatigue Research Center, University of Michigan, Ann Arbor, Michigan. yNeurology Department, University of Pennsylvania, Philadelphia, Pennsylvania. z Functional MRI Laboratory, University of Michigan, Ann Arbor, Michigan. xMGH/MIT/HMS Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, Massachusetts. {Department of Anesthesiology, Perioperative and Pain Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts. kDepartment of Radiology, Logan College of Chiropractic, Chesterfield, Missouri. **Department of Neurology, Bergmannsheil, Ruhr University Bochum, Bochum, Germany. yyDepartment of Psychology, University of Colorado Denver, Denver, Colorado. Abstract: The insular cortex (IC) and cingulate cortex (CC) are critically involved in pain perception. Previously we demonstrated that fibromyalgia (FM) patients have greater connectivity between the insula and default mode network at rest, and that changes in the degree of this connectivity were associated with changes in the intensity of ongoing clinical pain. In this study we more thoroughly evaluated the degree of resting-state connectivity to multiple regions of the IC in individuals with FM and healthy controls. We also investigated the relationship between connectivity, experimental pain, and current clinical chronic pain. Functional connectivity was assessed using resting-state functional magnetic resonance imaging in 18 FM patients and 18 age- and sex-matched healthy controls using predefined seed regions in the anterior, middle, and posterior IC. -

Cortical and Subcortical Mechanisms for Sound Processing Jennifer Blackwell University of Pennsylvania, [email protected]
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2019 Cortical And Subcortical Mechanisms For Sound Processing Jennifer Blackwell University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Neuroscience and Neurobiology Commons Recommended Citation Blackwell, Jennifer, "Cortical And Subcortical Mechanisms For Sound Processing" (2019). Publicly Accessible Penn Dissertations. 3275. https://repository.upenn.edu/edissertations/3275 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/3275 For more information, please contact [email protected]. Cortical And Subcortical Mechanisms For Sound Processing Abstract The uda itory cortex is essential for encoding complex and behaviorally relevant sounds. Many questions remain concerning whether and how distinct cortical neuronal subtypes shape and encode both simple and complex sound properties. In chapter 2, we tested how neurons in the auditory cortex encode water-like sounds perceived as natural by human listeners, but that we could precisely parametrize. The ts imuli exhibit scale-invariant statistics, specifically temporal modulation within spectral bands scaled with the center frequency of the band. We used chronically implanted tetrodes to record neuronal spiking in rat primary auditory cortex during exposure to our custom stimuli at different rates and cycle-decay constants. We found that, although neurons exhibited selectivity for subsets of stimuli with specific ts atistics, over the population responses were stable. These results contribute to our understanding of how auditory cortex processes natural sound statistics. In chapter 3, we review studies examining the role of different cortical inhibitory interneurons in shaping sound responses in auditory cortex. We identify the findings that support each other and the mechanisms that remain unexplored. -

Insular Cortex Corticotropin-Releasing Factor Integrates Stress Signaling with Social Decision Making
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.23.436680; this version posted March 23, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Insular cortex corticotropin releasing factor 1 Title: Insular cortex corticotropin-releasing factor integrates stress signaling with social decision making. 1 1 1 2 1 Authors: Nathaniel S. Rieger , Juan A. Varela , Alexandra Ng , Lauren Granata , Anthony Djerdjaj , Heather C. Brenhouse2, John P. Christianson1* 1Department of Psychology & Neuroscience, Boston College, 140 Commonwealth Ave, Chestnut Hill, MA 02467 USA. 2Psychology Department, Northeastern University, 360 Huntington Avenue, 115 Richards Hall, Boston, MA, 02115 *Corresponding Author: John Christianson, [email protected], 617-552-3970 Abstract Impairments in social cognition manifest in a variety of psychiatric disorders, making the neurobiological mechanisms underlying social decision making of particular translational importance. The insular cortex is consistently implicated in stress-related social and anxiety disorders, which are associated with diminished ability to make and use inferences about the emotions of others to guide behavior. We investigated how corticotropin releasing factor (CRF), a neuromodulator evoked by both self and social stressors, influenced the insula. In acute slices from male and female rats, CRF depolarized insular pyramidal neurons. In males, but not females, CRF suppressed presynaptic -

Brain Anatomy
BRAIN ANATOMY Adapted from Human Anatomy & Physiology by Marieb and Hoehn (9th ed.) The anatomy of the brain is often discussed in terms of either the embryonic scheme or the medical scheme. The embryonic scheme focuses on developmental pathways and names regions based on embryonic origins. The medical scheme focuses on the layout of the adult brain and names regions based on location and functionality. For this laboratory, we will consider the brain in terms of the medical scheme (Figure 1): Figure 1: General anatomy of the human brain Marieb & Hoehn (Human Anatomy and Physiology, 9th ed.) – Figure 12.2 CEREBRUM: Divided into two hemispheres, the cerebrum is the largest region of the human brain – the two hemispheres together account for ~ 85% of total brain mass. The cerebrum forms the superior part of the brain, covering and obscuring the diencephalon and brain stem similar to the way a mushroom cap covers the top of its stalk. Elevated ridges of tissue, called gyri (singular: gyrus), separated by shallow groves called sulci (singular: sulcus) mark nearly the entire surface of the cerebral hemispheres. Deeper groves, called fissures, separate large regions of the brain. Much of the cerebrum is involved in the processing of somatic sensory and motor information as well as all conscious thoughts and intellectual functions. The outer cortex of the cerebrum is composed of gray matter – billions of neuron cell bodies and unmyelinated axons arranged in six discrete layers. Although only 2 – 4 mm thick, this region accounts for ~ 40% of total brain mass. The inner region is composed of white matter – tracts of myelinated axons.