(Bursaphelenchus Xylophilus) Or Non-Pathogenic Nematodes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
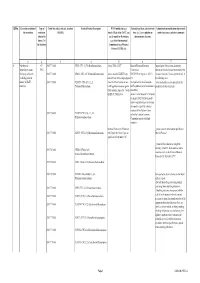
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -

Pines in the Arboretum
UNIVERSITY OF MINNESOTA MtJ ARBORETUM REVIEW No. 32-198 PETER C. MOE Pines in the Arboretum Pines are probably the best known of the conifers native to The genus Pinus is divided into hard and soft pines based on the northern hemisphere. They occur naturally from the up the hardness of wood, fundamental leaf anatomy, and other lands in the tropics to the limits of tree growth near the Arctic characteristics. The soft or white pines usually have needles in Circle and are widely grown throughout the world for timber clusters of five with one vascular bundle visible in cross sec and as ornamentals. In Minnesota we are limited by our cli tions. Most hard pines have needles in clusters of two or three mate to the more cold hardy species. This review will be with two vascular bundles visible in cross sections. For the limited to these hardy species, their cultivars, and a few hy discussion here, however, this natural division will be ignored brids that are being evaluated at the Arboretum. and an alphabetical listing of species will be used. Where neces Pines are readily distinguished from other common conifers sary for clarity, reference will be made to the proper groups by their needle-like leaves borne in clusters of two to five, of particular species. spirally arranged on the stem. Spruce (Picea) and fir (Abies), Of the more than 90 species of pine, the following 31 are or for example, bear single leaves spirally arranged. Larch (Larix) have been grown at the Arboretum. It should be noted that and true cedar (Cedrus) bear their leaves in a dense cluster of many of the following comments and recommendations are indefinite number, whereas juniper (Juniperus) and arborvitae based primarily on observations made at the University of (Thuja) and their related genera usually bear scalelikie or nee Minnesota Landscape Arboretum, and plant performance dlelike leaves that are opposite or borne in groups of three. -

Non-Expressway Master Plant List
MASTER PLANT LIST GENERAL INTRODUCTION TO PLANT LISTS Plants are living organisms. They possess variety in form, foliage and flower color, visual texture and ultimate size. There is variation in plants of the same species. Plants change: with seasons, with time and with the environment. Yet here is an attempt to categorize and catalogue a group of plants well suited for highway and expressway planting in Santa Clara County. This is possible because in all the existing variety of plants, there still remains a visual, morphological and taxonomical distinction among them. The following lists and identification cards emphasize these distinctions. 1 of 6 MASTER PLANT LIST TREES Acacia decurrens: Green wattle Acacia longifolia: Sydney golden wattle Acacia melanoxylon: Blackwood acacia Acer macrophyllum: Bigleaf maple Aesculus californica: California buckeye Aesculus carnea: Red horsechestnut Ailanthus altissima: Tree-of-heaven Albizia julibrissin: Silk tree Alnus cordata: Italian alder Alnus rhombifolia: White alder Arbutus menziesii: Madrone Calocedrus decurrens: Incense cedar Casuarina equisetifolia: Horsetail tree Casuarina stricta: Coast beefwood Catalpa speciosa: Western catalpa Cedrus deodara: Deodar cedar Ceratonia siliqua: Carob Cinnamomum camphora: Camphor Cordyline australis: Australian dracena Crataegus phaenopyrum: Washington thorn Cryptomeria japonica: Japanese redwood Cupressus glabra: Arizona cypress Cupressus macrocarpa: Monterey cypress Eriobotrya japonica: Loquat Eucalyptus camaldulensis: Red gum Eucalyptus citriodora: Lemon-scented -

CBD Fifth National Report
Fifth National Report of Japan to the Convention on Biological Diversity Government of Japan March 2014 Contents Executive Summary 1 Chapter 1 Biodiversity: the current situation, trends and threats 7 1.1 Importance of biodiversity 7 (1) Characteristics of biodiversity in Japan from the global perspective 7 (2) Biodiversity that supports life and livelihoods 9 (3) Japan causing impacts on global biodiversity 10 (4) The economic valuation of biodiversity 11 1.2 Major changes to the biodiversity situation and trends 12 (1) The current situation of ecosystems 12 (2) The current situation of threatened wildlife 17 (3) Impacts of the Great East Japan Earthquake on biodiversity 19 1.3 The structure of the biodiversity crisis 21 (1) The four crises of biodiversity 21 (2) Japan Biodiversity Outlook (JBO) 22 1.4 The impacts of changes in biodiversity on ecosystem services, socio-economy, and culture 24 (1) Changes in the distribution of medium and large mammals and the expansion of conflicts 24 (2) Alien species 24 (3) Impacts of changes in the global environment on biodiversity 26 1.5 Future scenarios for biodiversity 28 (1) Impacts of the global warming 28 (2) The impacts of ocean acidification on coral reefs 29 (3) The forecasted expansion in the distribution of sika deer (Cervus nippon ) 30 (4) Second crisis (caused by reduced human activities) 30 Chapter 2 Implementation of the National Biodiversity Strategy and Mainstreaming Biodiversity 32 2.1 Background to the formulation of the National Biodiversity Strategy of Japan and its development -

Pinus Densiflora 'Pendula'1
Fact Sheet FPS-481 October, 1999 Pinus densiflora ‘Pendula’1 Edward F. Gilman2 Introduction Growth rate: slow Texture: fine Japanese Red Pine reaches a height and spread of 30 to 50 Foliage feet in the landscape growing much taller in the woods. Needles are arranged in pairs and remain on the tree for about Leaf arrangement: spiral three years. A distinguishing feature of this tree is the often Leaf type: simple crooked or sweeping trunk which shows reddish-orange peeling Leaf margin: entire bark. Because lower branches are held nearly horizontal on the trunk forming a picturesque silhouette in the landscape it is used Leaf shape: needle-like (filiform) best as a specimen, not as a mass planting. Needles may turn Leaf venation: parallel yellowish during winter on some soils. Leaf type and persistence: evergreen Leaf blade length: 4 to 8 inches Leaf color: green General Information Fall color: no fall color change Fall characteristic: not showy Scientific name: Pinus densiflora ‘Pendula’ Pronunciation: PYE-nuss den-siff-FLOR-ruh Flower Common name(s): Weeping Japanese Red Pine Family: Pinaceae Flower color: yellow Plant type: tree Flower characteristic: inconspicuous and not showy USDA hardiness zones: 3B through 7A (Fig. 1) Planting month for zone 7: year round Fruit Origin: not native to North America Uses: bonsai Fruit shape: oval Availablity: grown in small quantities by a small number of Fruit length: 1 to 3 inches nurseries Fruit cover: dry or hard Fruit color: tan Fruit characteristic: persists on the plant Description Height: 6 to 10 feet Trunk and Branches Spread: 10 to 15 feet Plant habit: weeping; spreading Trunk/bark/branches: typically multi-trunked or clumping Plant density: dense stems; no thorns 1.This document is Fact Sheet FPS-481, one of a series of the Environmental Horticulture Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. -

Redalyc.Catalogue of Eucosmini from China (Lepidoptera: Tortricidae)
SHILAP Revista de Lepidopterología ISSN: 0300-5267 [email protected] Sociedad Hispano-Luso-Americana de Lepidopterología España Zhang, A. H.; Li, H. H. Catalogue of Eucosmini from China (Lepidoptera: Tortricidae) SHILAP Revista de Lepidopterología, vol. 33, núm. 131, septiembre, 2005, pp. 265-298 Sociedad Hispano-Luso-Americana de Lepidopterología Madrid, España Available in: http://www.redalyc.org/articulo.oa?id=45513105 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative 265 Catalogue of Eucosmini from 9/9/77 12:40 Página 265 SHILAP Revta. lepid., 33 (131), 2005: 265-298 SRLPEF ISSN:0300-5267 Catalogue of Eucosmini from China1 (Lepidoptera: Tortricidae) A. H. Zhang & H. H. Li Abstract A total of 231 valid species in 34 genera of Eucosmini (Lepidoptera: Tortricidae) are included in this catalo- gue. One new synonym, Zeiraphera hohuanshana Kawabe, 1986 syn. n. = Zeiraphera thymelopa (Meyrick, 1936) is established. 28 species are firstly recorded for China. KEY WORDS: Lepidoptera, Tortricidae, Eucosmini, Catalogue, new synonym, China. Catálogo de los Eucosmini de China (Lepidoptera: Tortricidae) Resumen Se incluyen en este Catálogo un total de 233 especies válidas en 34 géneros de Eucosmini (Lepidoptera: Tor- tricidae). Se establece una nueva sinonimia Zeiraphera hohuanshana Kawabe, 1986 syn. n. = Zeiraphera thymelopa (Meyrick, 1938). 28 especies se citan por primera vez para China. PALABRAS CLAVE: Lepidoptera, Tortricidae, Eucosmini, catálogo, nueva sinonimia, China. Introduction Eucosmini is the second largest tribe of Olethreutinae in Tortricidae, with about 1000 named spe- cies in the world (HORAK, 1999). -

Pine - Wikipedia Visited on 06/20/2017
Pine - Wikipedia Visited on 06/20/2017 Not logged in Talk Contributions Create account Log in Article Talk Read Edit View history Pine From Wikipedia, the free encyclopedia Main page Contents This article is about the tree. For other uses of the term "pine", see Pine (disambiguation). Featured content A pine is any conifer in the genus Pinus, /ˈpiːnuːs/,[1] of the Current events Pine tree family Pinaceae . Pinus is the sole genus in the subfamily Random article Pinoideae. The Plant List compiled by the Royal Botanic Donate to Wikipedia Wikipedia store Gardens, Kew and Missouri Botanical Garden accepts 126 species names of pines as current, together with 35 unresolved Interaction species and many more synonyms.[2] Help About Wikipedia Contents [hide] Community portal 1 Etymology Recent changes 2 Taxonomy, nomenclature and codification Contact page 3 Distribution 4 Description Tools 4.1 Foliage What links here 4.2 Cones Related changes 5 Ecology Upload file 6 Use Special pages 6.1 Farming Permanent link Japanese red pine (Pinus densiflora), Page information 6.2 Food North Korea Wikidata item 6.3 Folk Medicine Cite this page 7 See also Scientific classification 8 Notes Kingdom: Plantae Print/export 9 References Division: Pinophyta Create a book 10 Bibliography Class: Pinopsida Download as PDF 11 External links Printable version Order: Pinales In other projects Family: Pinaceae Etymology [edit] Wikimedia Commons Genus: Pinus Wikispecies The modern L. English name pine Subgenera Languages derives from Latin Subgenus Strobus Afrikaans pinus, which some Alemannisch Subgenus Pinus have traced to the Indo-European See Pinus classification for Ænglisc complete taxonomy to species ’base *pīt- ‘resin العربية level. -

Breeding and Genetic Resources of Five-Needle Pines
Estimation of Heritabilities and Clonal Contribution Based on the Flowering Assessment in Two Clone Banks of Pinus koraiensis Sieb. et Zucc. Wan-Yong Choi Kyu-Suk Kang Sang-Urk Han Seong-Doo Hur Abstract—Reproductive characteristics of 161 Korean pine (Pinus the northeastern part of Eurasia. It usually occurs as a koraiensis Sieb. et Zucc.) clones were surveyed at two clone banks for mixed forest stand consisting of various broad-leaved tree 3 years. These clone banks were established at Yongin and Chunchon species and other conifers. Korean pine has been widely (mid-Korea) in 1983. Characteristics in female and male strobili planted as a pure stand accounting for about 30 percent of were spatial (between locations) and temporal (among investigated the yearly planting areas in Korea due to its high-quality times) variables. Broad sense heritabilities were found to vary timber and edible seeds. A breeding program for this species between 0.20 - 0.46 in females and between 0.34 to 0.56 in males. has been conducted since 1959 and has resulted in the Among 161 clones, 32 clones (20 percent of the total clones) ac- establishment of 98 ha seed orchards (Mirov 1967, Chun counted for 42 to 54 percent of clonal contribution in female strobili 1992, Choi 1993, Wang 2001). and 83 to 96 percent in male strobili, suggesting that the clonal The main goal for seed orchards is large-scale production contribution for male parents was severely unbalanced compared to of genetically improved seeds that maintain genetic diver- that for female parents. The effective population numbers varied sity to prevent inbreeding depression. -

Rutgers Home Gardeners School: Conifers Plant List
Conifers For The Garden By Walter Cullerton 1 Abies alba 'Pyramadalis' (White fir) 2 Abies balsamea 'Hudsonia' (Balsam fir) 3 Abies koreana 'Horstmann Silberlocke' X3 (Korean Fir) 4 Abies koreana 'Green Carpet' (Korean Fir) 5 Abies koreana 'Blauwe Zwo' (Korean Fir) 6 Abies kpreana 'Ice Breaker' (Korean Fir) 7 Abies fraseri 'Stricta' (Fraser Fir) 8 Abies procera 'Silver' (Noble Fir) 9 Abies lasiocarpa 'Compacta' (Subalpine Fir) The Cedars 10 Cedrus deodara Aurea? (Himalayan Cedar) 11 Cedrus libanii cones (Cedar of Lebanon) 12 Cedrus atlantica 'Glauca' (Atlantic Cedar or Blue Atlas) 13 Cedrus atlantica 'Fastigiata 14 Cedrus atlantica 'Glauca Pendula' The Plum Yew 15 Cephalotaxus harringtonia 'Prostrata' (Plum Yew) 16 Cephalotaxus harringtonia 'Kelly's Gold' The False Cedars 17 Chamaecyparis nookatensis 'Green Arrow' X2 (Alaskan Cedar) 18 Chamaecyparis nookatensis 'Van Den Aker' 19 Chamaecyparis obtusa 'Lynn's Golden' (Hinoki Cypress) 20 Chamaecyparis obtusa Fernspray Gold' 21 Chamaecyparis obtusa 'Crippsii' 22 Chamaecyparis pisifera 'Snow' (Sawara Cypress) 23 Chamaecyparis pisifera 'Variegata' The Japanese Cedar 24 Cryptomeria japonica 'Gyokurya' (Japanese Cedar) Cryptomeria japonica 'Spiralis' (Japanese Cedar) The China Fir 25 Cunninghamia lanceolata 'Glauca' 26 Cunninghamia lanceolata needles Arizona Cypress 27 Cupressus arizonica var glabra 'Blue Ice' (Arizona Cypress) The Junipers 28 Juniperus communis 'Gold Cone' (Common Juniper) 29 Jumiperus rigida 'Pendula' (Temple Juniper) 30 Juniperus viginiana 'Gray Owl' a scene (Eastern Red Cedar) -

Download the Poster Presentation List Here (4/20/2018)
Poster presentations at the 8th EAFES (P1: 21 April, P2: 22 April) Poster First Name Family Name Title ID of 1st author of 1st author Relationship between floral tube length and proboscis length in nocturnal P1-1 Kota Sakagami moths: Do long-tongued moths visit long-tubed flowers? Ectomycorrhizal fungi in Nansei Islands; The comparison between P1-2 Ryo Yasui Takashima-Islands and Hikushima-Islands Natural hybridization between Japanese two nominal medaka species P1-3 Yuka Iguchi observed in the Yura River basin, Japan Equal size cell divisions during gametogenesis in the marine green alga, P1-4 Yusuke Horinouchi Monostroma angicava. The microbial community structures of surface soil and organic layer of P1-5 Masataka Nakayama three different types of forests in northeast of Japan The spatial genetic structure of mountainous populations in endangered P1-6 Naohiro Ishii gentian Gentiana yakushimensis Height-related changes in hydraulic structure of old Cinnamomum P1-7 Yuiko Noguchi camphora: tradeoff between hydraulic conductivity and safety Tracking and Acoustic Luring of the Ryukyu Tube-nosed Bat Murina P1-8 Jason H. Preble ryukyuana P1-9 Keitaro Umemura Hybridization between endangered bitterling species in natural water area. Does phenotypic plasticity in the leaves of host plants promote resource P1-10 Haruna Ohsaki partition between leaf beetles? Are stray Tibetan mastiffs a potential threat to snow leopards on the P1-11 Mingyu Liu Tibetan Plateau? The soil prokaryotic community under halophytic tamarisk shrubs P1-12 Chikae Iwaoka -

Korean Red List of Threatened Species Korean Red List Second Edition of Threatened Species Second Edition Korean Red List of Threatened Species Second Edition
Korean Red List Government Publications Registration Number : 11-1480592-000718-01 of Threatened Species Korean Red List of Threatened Species Korean Red List Second Edition of Threatened Species Second Edition Korean Red List of Threatened Species Second Edition 2014 NIBR National Institute of Biological Resources Publisher : National Institute of Biological Resources Editor in President : Sang-Bae Kim Edited by : Min-Hwan Suh, Byoung-Yoon Lee, Seung Tae Kim, Chan-Ho Park, Hyun-Kyoung Oh, Hee-Young Kim, Joon-Ho Lee, Sue Yeon Lee Copyright @ National Institute of Biological Resources, 2014. All rights reserved, First published August 2014 Printed by Jisungsa Government Publications Registration Number : 11-1480592-000718-01 ISBN Number : 9788968111037 93400 Korean Red List of Threatened Species Second Edition 2014 Regional Red List Committee in Korea Co-chair of the Committee Dr. Suh, Young Bae, Seoul National University Dr. Kim, Yong Jin, National Institute of Biological Resources Members of the Committee Dr. Bae, Yeon Jae, Korea University Dr. Bang, In-Chul, Soonchunhyang University Dr. Chae, Byung Soo, National Park Research Institute Dr. Cho, Sam-Rae, Kongju National University Dr. Cho, Young Bok, National History Museum of Hannam University Dr. Choi, Kee-Ryong, University of Ulsan Dr. Choi, Kwang Sik, Jeju National University Dr. Choi, Sei-Woong, Mokpo National University Dr. Choi, Young Gun, Yeongwol Cave Eco-Museum Ms. Chung, Sun Hwa, Ministry of Environment Dr. Hahn, Sang-Hun, National Institute of Biological Resourses Dr. Han, Ho-Yeon, Yonsei University Dr. Kim, Hyung Seop, Gangneung-Wonju National University Dr. Kim, Jong-Bum, Korea-PacificAmphibians-Reptiles Institute Dr. Kim, Seung-Tae, Seoul National University Dr. -

1688-1701 Issn 2077-4605
Middle East Journal of Agriculture Volume : 07 | Issue : 04 | Oct.-Dec. | 2018 Research Pages:1688-1701 ISSN 2077-4605 Taxonomic revision of Piuus L. in Egypt Fatema S. Mohamed Flora and Phytotaxonomy Researches Department, Horticulture Research Institute, ARC., Giza, Egypt. Received: 20 Oct. 2018 / Accepted 10 Dec. 2018 / Publication date: 24 Dec. 2018 ABSTRACT This study includes taxonomic revision, documentation, description and distribution of Pinus species cultivated in Egypt. Fresh specimens were collected from three gardens; The Agricultural Museum, El-Orman and the Zoo Gardens. Seven species and one variety were collected, identified, described and compared with the specimens in the herbarium of Flora and Phytotaxonomy Researches Department (CAIM). Three new species and one variety were deposited in the herbarium namely; P. brutia Ten., P. brutia Ten. var. eldarica (Medw.) Silba., P. canariensis C. Smith and P. sylvestris L. Herbarium specimens were examined and photographed to help in establishing database information and to be available for the taxonomic researchers to further study of this genus. This study showed the importance of morphological traits for taxonomic evaluation among the studied taxa of Pinus in Egypt. Moreover a constructed key to Pinus species is provided. Keywords: Pinaceae, Pinus, Resinous trees, Coniferous Introduction The family Pinaceae is trees or shrubs, including many of the well-known conifers of commercial importance contains 11 genera and 220 species (Farjon 1998). According to the characters of cone and seed, Pinaceae are divided into four subfamilies; Pinoideae, Piceoideae, Laricoideae and Abietoideae (Frankis 1989 and Farjon 1990). Among Coniferous the pines constitute by far the most important group regarding either from the point of view of number of species or that of economic value (Bean 1922).