Gene Expression Profiling of Liver and Mammary Tissues of Lactating Dairy Cows
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Role of Alpha Oxidation in Lipid Metabolism, 2017
THE ROLE OF ALPHA OXIDATION IN LIPID METABOLISM Benjamin John Jenkins Darwin College Medical Research Council – Human Nutrition Research Department of Biochemistry University of Cambridge This dissertation is submitted for the degree of Doctor of Philosophy July 2018 DECLARATION This dissertation is the result of my own work and includes nothing, which is the outcome of work done in collaboration except as declared in the preface and specified in the text. It is not substantially the same as any that I have submitted, or, is being concurrently submitted for a degree or diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the Preface and specified in the text. I further state that no substantial part of my dissertation has already been submitted, or, is being concurrently submitted for any such degree, diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the Preface and specified in the text. In accordance with the University of Cambridge guidelines, this thesis does not exceed 60,000 words. Signed: ______________________________________________________________ Date: _______________________________________________________________ Benjamin John Jenkins BSc. MSc. Darwin College, Silver Street, Cambridge, CB3 9EU by i 2014 Word Template Template Friedman Friedman & Morgan Morgan & ii The Role of Alpha Oxidation in Lipid Metabolism, 2017 ABSTRACT Recent findings have shown an inverse association between the circulating levels of pentadecanoic acid (C15:0) and heptadecanoic acid (C17:0) with the risk of pathological development in type 2 diabetes, cardio vascular disease and neurological disorders. From previously published research, it has been said that both these odd chain fatty acids are biomarkers of their dietary intake and are significantly correlated to dietary ruminant fat intake. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Dataset for the Quantitative Proteomics Analysis of the Primary Hepatocellular Carcinoma with Single and Multiple Lesions
Data in Brief 5 (2015) 226–240 Contents lists available at ScienceDirect Data in Brief journal homepage: www.elsevier.com/locate/dib Data Article Dataset for the quantitative proteomics analysis of the primary hepatocellular carcinoma with single and multiple lesions Xiaohua Xing a,b, Yao Huang c, Sen Wang a,b, Minhui Chi a,b,c, Yongyi Zeng a,b,c, Lihong Chen a,b,c, Ling Li a,b,c, Jinhua Zeng a,b,c, Minjie Lin a,b, Xiao Han d, Jingfeng Liu a,b,c, Xiaolong Liu a,b,n a The Liver Center of Fujian Province, Fujian Medical University, Fuzhou 350025, People’s Republic of China b The United Innovation of Mengchao Hepatobiliary Technology Key Laboratory of Fujian Province, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou 350025, People’s Republic of China c Liver Disease Center, The First Affiliated Hospital of Fujian Medical University, Fuzhou 350007, People’s Republic of China d Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, People’s Republic of China article info abstract Article history: Hepatocellular Carcinoma (HCC) is one of the most common malig- Received 18 August 2015 nant tumor, which is causing the second leading cancer-related death Received in revised form worldwide. The tumor tissues and the adjacent noncancerous tissues 27 August 2015 obtained from HCC patients with single and multiple lesions were Accepted 28 August 2015 quantified using iTRAQ. A total of 5513 proteins (FDR of 1%) were Available online 8 September 2015 identified which correspond to roughly 27% of the total liver proteome. -

Fatty Acid Oxidation
FATTY ACID OXIDATION 1 FATTY ACIDS A fatty acid contains a long hydrocarbon chain and a terminal carboxylate group. The hydrocarbon chain may be saturated (with no double bond) or may be unsaturated (containing double bond). Fatty acids can be obtained from- Diet Adipolysis De novo synthesis 2 FUNCTIONS OF FATTY ACIDS Fatty acids have four major physiological roles. 1)Fatty acids are building blocks of phospholipids and glycolipids. 2)Many proteins are modified by the covalent attachment of fatty acids, which target them to membrane locations 3)Fatty acids are fuel molecules. They are stored as triacylglycerols. Fatty acids mobilized from triacylglycerols are oxidized to meet the energy needs of a cell or organism. 4)Fatty acid derivatives serve as hormones and intracellular messengers e.g. steroids, sex hormones and prostaglandins. 3 TRIGLYCERIDES Triglycerides are a highly concentrated stores of energy because they are reduced and anhydrous. The yield from the complete oxidation of fatty acids is about 9 kcal g-1 (38 kJ g-1) Triacylglycerols are nonpolar, and are stored in a nearly anhydrous form, whereas much more polar proteins and carbohydrates are more highly 4 TRIGLYCERIDES V/S GLYCOGEN A gram of nearly anhydrous fat stores more than six times as much energy as a gram of hydrated glycogen, which is likely the reason that triacylglycerols rather than glycogen were selected in evolution as the major energy reservoir. The glycogen and glucose stores provide enough energy to sustain biological function for about 24 hours, whereas the Triacylglycerol stores allow survival for several weeks. 5 PROVISION OF DIETARY FATTY ACIDS Most lipids are ingested in the form of triacylglycerols, that must be degraded to fatty acids for absorption across the intestinal epithelium. -

Epistasis-Driven Identification of SLC25A51 As a Regulator of Human
ARTICLE https://doi.org/10.1038/s41467-020-19871-x OPEN Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import Enrico Girardi 1, Gennaro Agrimi 2, Ulrich Goldmann 1, Giuseppe Fiume1, Sabrina Lindinger1, Vitaly Sedlyarov1, Ismet Srndic1, Bettina Gürtl1, Benedikt Agerer 1, Felix Kartnig1, Pasquale Scarcia 2, Maria Antonietta Di Noia2, Eva Liñeiro1, Manuele Rebsamen1, Tabea Wiedmer 1, Andreas Bergthaler1, ✉ Luigi Palmieri2,3 & Giulio Superti-Furga 1,4 1234567890():,; About a thousand genes in the human genome encode for membrane transporters. Among these, several solute carrier proteins (SLCs), representing the largest group of transporters, are still orphan and lack functional characterization. We reasoned that assessing genetic interactions among SLCs may be an efficient way to obtain functional information allowing their deorphanization. Here we describe a network of strong genetic interactions indicating a contribution to mitochondrial respiration and redox metabolism for SLC25A51/MCART1, an uncharacterized member of the SLC25 family of transporters. Through a combination of metabolomics, genomics and genetics approaches, we demonstrate a role for SLC25A51 as enabler of mitochondrial import of NAD, showcasing the potential of genetic interaction- driven functional gene deorphanization. 1 CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria. 2 Laboratory of Biochemistry and Molecular Biology, Department of Biosciences, Biotechnologies and Biopharmaceutics, -

Placental Transcriptome Adaptations to Maternal Nutrient Restriction in Sheep
International Journal of Molecular Sciences Article Placental Transcriptome Adaptations to Maternal Nutrient Restriction in Sheep Chelsie B. Steinhauser 1,†, Colleen A. Lambo 2,†, Katharine Askelson 1, Gregory W. Burns 3, Susanta K. Behura 4,5, Thomas E. Spencer 4, Fuller W. Bazer 1 and Michael Carey Satterfield 1,* 1 Department of Animal Science, Texas A & M University, College Station, TX 77843, USA; [email protected] (C.B.S.); [email protected] (K.A.); [email protected] (F.W.B.) 2 Department of Veterinary Physiology and Pharmacology, Texas A & M University, College Station, TX 77843, USA; [email protected] 3 Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI 49503, USA; [email protected] 4 Division of Animal Sciences, University of Missouri, Columbia, MO 65211, USA; [email protected] (S.K.B.); [email protected] (T.E.S.) 5 Institute for Data Science and Informatics, University of Missouri, Columbia, MO 65211, USA * Correspondence: csatterfi[email protected]; Tel.: +1-979-845-6448 † These authors contributed equally to this work. Abstract: Placental development is modified in response to maternal nutrient restriction (NR), resulting in a spectrum of fetal growth rates. Pregnant sheep carrying singleton fetuses and fed either 100% (n = 8) or 50% (NR; n = 28) of their National Research Council (NRC) recommended intake from days 35–135 of pregnancy were used to elucidate placentome transcriptome alterations at both Citation: Steinhauser, C.B.; Lambo, day 70 and day 135. NR fetuses were further designated into upper (NR NonSGA; n = 7) and lower C.A.; Askelson, K.; Burns, G.W.; quartiles (NR SGA; n = 7) based on day 135 fetal weight. -

Hepatic Proteome Analysis Reveals Altered Mitochondrial Metabolism and Suppressed Acyl-Coa Synthetase-1 in Colon-26 Tumor-Induce
UCLA UCLA Previously Published Works Title Hepatic proteome analysis reveals altered mitochondrial metabolism and suppressed acyl- CoA synthetase-1 in colon-26 tumor-induced cachexia. Permalink https://escholarship.org/uc/item/5xb4k7mj Journal Physiological genomics, 52(5) ISSN 1094-8341 Authors Khamoui, Andy V Tokmina-Roszyk, Dorota Rossiter, Harry B et al. Publication Date 2020-05-01 DOI 10.1152/physiolgenomics.00124.2019 Peer reviewed eScholarship.org Powered by the California Digital Library University of California 1 Hepatic proteome analysis reveals altered mitochondrial metabolism and suppressed acyl-CoA 2 synthetase-1 in colon-26 tumor-induced cachexia 3 4 Andy V. Khamoui1,2*, Dorota Tokmina-Roszyk2,3, Harry B. Rossiter5,6, Gregg B. Fields2,3,4, Nishant P. 5 Visavadiya1 6 7 1Department of Exercise Science and Health Promotion, Florida Atlantic University, Boca Raton, FL, USA; 8 2Institute for Human Health & Disease Intervention, Florida Atlantic University, Boca Raton, FL, USA; 9 3Department of Chemistry & Biochemistry, Florida Atlantic University, Jupiter, FL, USA; 4Department of 10 Chemistry, The Scripps Research Institute, Jupiter, FL, USA; 5Division of Respiratory and Critical Care 11 Physiology and Medicine, Department of Medicine, The Lundquist Institute for Biomedical Innovation at 12 Harbor-UCLA Medical Center, Torrance, CA, USA; 6Faculty of Biological Sciences, University of Leeds, Leeds, 13 UK 14 15 Running Title: Hepatic proteome in cancer cachexia 16 17 *Correspondence 18 Andy V. Khamoui PhD 19 Department of Exercise Science and Health Promotion 20 Florida Atlantic University 21 777 Glades Rd, FH-11A, Rm 128-B 22 Boca Raton, FL 33431 23 Tel: (561) 297-4450 | Email: [email protected] 24 25 26 27 28 1 Downloaded from journals.physiology.org/journal/physiolgenomics by Harry Rossiter (047.144.146.194) on March 14, 2020. -

The Inverse Agonist DG172 Triggers a Pparβ/Δ-Independent Myeloid Lineage Shift and Promotes GM-CSF/IL-4-Induced Dendritic Cell Differentiation
Downloaded from molpharm.aspetjournals.org at ASPET Journals on September 26, 2021 1 MOL #94672 #94672 MOL -independent myeloid lineage shift and -independent myeloid lineage δ / β nagel, Wolfgang Meissner, Gavin Giehl, Cornelia Giehl, Cornelia Gavin Wolfgang Meissner, nagel, This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. -
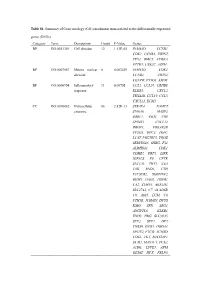
Table S1. Summary of Gene Ontology (GO) Enrichment Terms Related To
Table S1. Summary of Gene ontology (GO) enrichment terms related to the differentially expressed genes (DEGs) Category Term Description Count P-Value Genes BP GO:0051301 Cell division 12 1.13E-04 FAM83D, CCNB1, CDK1, CCNB2, ZWINT, TPX2, BIRC5, AURKA, PTTG1, UBE2C, ASPM BP GO:0007067 Mitotic nuclear 8 0.003429 FAM83D, CDK1, division CCNB2, ZWINT, CENPW, PTTG1, ASPM BP GO:0006954 Inflammatory 11 0.00705 CCL2, CCL20, CRHBP, response KLKB1, CXCL2, TBXA2R, CCL19, CCL5, CXCL12, ECM1 CC GO:0070062 Extracellular 86 2.82E-13 STEAP4, NAMPT, exosome S100A8, MASP2, SORL1, ANO1, CRP, SPINK1, CXCL12, BBOX1, PRKAR2B, TP53I3, GPC3, TKFC, LCAT, PGLYRP2, PROZ, SERPINA4, SHBG, F11, ALDH6A1, CDK1, CDHR2, FBP1, LIFR, IGFALS, F9, CFTR, SLC3A1, THY1, C8A, C8B, RND3, CTH, TACSTD2, SERPINF2, BHMT, HAO2, HSPB1, CA2, STMN1, AKR1D1, SLC27A2, C7, ACADSB, C9, ASS1, LUM, C6, PTH1R, JCHAIN, DPYS, KMO, SFN, ARG1, ANGPTL6, KLKB1, ENO3, HRG, SLC39A5, SPP2, SPP1, DPT, THBS4, GPD1, HOGA1, EPHX2, FTCD, ACMSD, CD5L, TKT, RACGAP1, ECM1, MAN1C1, PCK1, A1BG, LYVE1, AFM, GLYAT, HPX, FBLN5, PON1, CP, HPGD, IGFBP3, FABP5 CC GO:0072562 Blood 13 7.61E-09 C9, JCHAIN, CD5L, microparticle C8A, A1BG, AFM, BCHE, HPX, SERPINF2, APOA5, PON1, HRG, CP CC GO:0005615 Extracellular 39 2.25E-06 XDH, MBL2, CCL2, space S100A8, ADAMTS13, CRHBP, LUM, CRP, CXCL2, SORL1, CD109, SPINK1, CCL5, PTGIS, CCL20, BCHE, ANG, KLKB1, PROZ, SERPINA4, ENO3, NRG1, SPP1, THBS4, DPT, F11, IGF1, CCL19, F9, DBH, ECM1, BMPER, SERPINF2, TACSTD2, FBLN5, PON1, HSPB1, IGFBP3 CC GO:0005739 Mitochondrion 29 0.002339 ETNPPL, -

Integrated Physiology and Systems Biology of Ppara
Review Integrated physiology and systems biology of PPARa Sander Kersten* ABSTRACT The Peroxisome Proliferator Activated Receptor alpha (PPARa) is a transcription factor that plays a major role in metabolic regulation. This review addresses the functional role of PPARa in intermediary metabolism and provides a detailed overview of metabolic genes targeted by PPARa, with a focus on liver. A distinction is made between the impact of PPARa on metabolism upon physiological, pharmacological, and nutritional activation. Low and high throughput gene expression analyses have allowed the creation of a comprehensive map illustrating the role of PPARa as master regulator of lipid metabolism via regulation of numerous genes. The map puts PPARa at the center of a regulatory hub impacting fatty acid uptake, fatty acid activation, intracellular fatty acid binding, mitochondrial and peroxisomal fatty acid oxidation, ketogenesis, triglyceride turnover, lipid droplet biology, gluconeogenesis, and bile synthesis/secretion. In addition, PPARa governs the expression of several secreted proteins that exert local and endocrine functions. Ó 2014 The Author. Published by Elsevier GmbH. This is an open access article under the CC BY-NC-SA license (http://creativecommons.org/licenses/by-nc-sa/3.0/). Keywords PPARa; Liver; Transcriptional networks; Lipid metabolism; Expression profiling; Metabolic homeostasis; Systems biology 1. INTRODUCTION deacetylase and histone acetyltransferase activity, respectively, necessary for the assembly of the transcription initiation complex [10]. PPARa was the first member to be cloned of a small subfamily of Readers are referred to another review for more detailed information nuclear receptors called Peroxisome Proliferators Activated Receptors on co-activators in PPAR-dependent gene regulation [11]. -

Fatty Acid Oxidation- Notes
Fatty acid oxidation- Notes See how fatty acids are broken down and used to generate ATP . Fatty acids provide highly efficient energy storage, delivering more energy per gram than carbohydrates like glucose. In tissues with high energy requirement, such as heart, up to 50– 70% of energy, in the form of ATP production, comes from fatty acid (FA) beta-oxidation. During fatty acid β-oxidation long chain acyl-CoA molecules – the main components of FAs – are broken to acetyl-CoA molecules. Fatty acid transport into mitochondria Fatty acids are activated for degradation by conjugation with coenzyme A (CoA) in the cytosol. The long-chain fatty-acyl-CoA is then modified by carnitine palmitoyltransferase 1 (CPT1) to acylcarnitine and transported across the inner mitochondrial membrane by carnitine translocase (CAT). CPT2 then coverts the long chain acylcarnitine back to long- chain acyl-CoA before beta-oxidation. Breakdown of fats yields fatty acids and glycerol. Glycerol can be readily converted to DHAP for oxidation in glycolysis or synthesis into glucose in gluconeogenesis. Fatty acids are broken down in two carbon units of acetyl-CoA. To be oxidized, they must be transported through the cytoplasm attached to coenzyme A and moved into mitochondria. The latter step requires removal of the CoA and attachment of the fatty acid to a molecule of carnitine. The carnitine complex is transported across the inner membrane of the mitochondrion after which the fatty acid is reattached to coenzyme A in the mitochondrial matrix. Dr Anjali Saxena Page 1 Figure- Movement of Acyl-CoAs into the Mitochondrial Matrix The process of fatty acid oxidation, called beta oxidation, is fairly simple.