View Using BLAT Program [6] with Default Parameters
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Efficacy and Mechanistic Evaluation of Tic10, a Novel Antitumor Agent
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2012 Efficacy and Mechanisticv E aluation of Tic10, A Novel Antitumor Agent Joshua Edward Allen University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Oncology Commons Recommended Citation Allen, Joshua Edward, "Efficacy and Mechanisticv E aluation of Tic10, A Novel Antitumor Agent" (2012). Publicly Accessible Penn Dissertations. 488. https://repository.upenn.edu/edissertations/488 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/488 For more information, please contact [email protected]. Efficacy and Mechanisticv E aluation of Tic10, A Novel Antitumor Agent Abstract TNF-related apoptosis-inducing ligand (TRAIL; Apo2L) is an endogenous protein that selectively induces apoptosis in cancer cells and is a critical effector in the immune surveillance of cancer. Recombinant TRAIL and TRAIL-agonist antibodies are in clinical trials for the treatment of solid malignancies due to the cancer-specific cytotoxicity of TRAIL. Recombinant TRAIL has a short serum half-life and both recombinant TRAIL and TRAIL receptor agonist antibodies have a limited capacity to perfuse to tissue compartments such as the brain, limiting their efficacy in certain malignancies. To overcome such limitations, we searched for small molecules capable of inducing the TRAIL gene using a high throughput luciferase reporter gene assay. We selected TRAIL-inducing compound 10 (TIC10) for further study based on its induction of TRAIL at the cell surface and its promising therapeutic index. TIC10 is a potent, stable, and orally active antitumor agent that crosses the blood-brain barrier and transcriptionally induces TRAIL and TRAIL-mediated cell death in a p53-independent manner. -

(12) Patent Application Publication (10) Pub. No.: US 2009/0297536A1 Chin Et Al
US 20090297536A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0297536A1 Chin et al. (43) Pub. Date: Dec. 3, 2009 (54) COMPOSITIONS, KITS, AND METHODS FOR Related U.S. Application Data IDENTIFICATION, ASSESSMENT, (60) Provisional application No. 60/575,795, filed on May PREVENTION AND THERAPY OF CANCER 28, 2004, provisional application No. 60/580,337, filed on Jun. 15, 2004. (75) Inventors: Lynda Chin, Brookline, MA (US); Publication Classification Cameron W. Brennan, New York, NY (US); Ronald A. DePinho, (51) Int. Cl. Brookline, MA (US); Andrew J. A 6LX 39/395 (2006.01) Aguirre, Boston, MA (US) CI2O I/68 (2006.01) C40B 30/00 (2006.01) AOIK 67/00 (2006.01) Correspondence Address: A 6LX 3L/7052 (2006.01) FOLEY HOAG, LLP A638/02 (2006.01) PATENT GROUP, WORLD TRADE CENTER A638/16 (2006.01) WEST C07K I4/00 (2006.01) 155 SEAPORT BLVD C7H 2L/00 (2006.01) BOSTON, MA 02110 (US) C07K 6/00 (2006.01) A6IP35/00 (2006.01) (73) Assignee: Dana-Farber Cancer Institute, (52) U.S. Cl. ................ 424/172.1; 435/6:506/7; 800/10: Inc., Boston, MA (US) 424/183.1; 424/178.1: 514/44 A: 514/2: 514/12: 530/350:536/23.1; 530/389.1 (21) Appl. No.: 11/597,825 (57) ABSTRACT The invention relates to compositions, kits, and methods for (22) PCT Filed: May 27, 2005 detecting, characterizing, preventing, and treating human cancer. A variety of chromosomal regions (MCRs) and mark (86). PCT No.: PCT/US05/18850 ers corresponding thereto, are provided, wherein alterations in the copy number of one or more of the MCRs and/or S371 (c)(1), alterations in the amount, structure, and/or activity of one or (2), (4) Date: Jun. -

(P -Value<0.05, Fold Change≥1.4), 4 Vs. 0 Gy Irradiation
Table S1: Significant differentially expressed genes (P -Value<0.05, Fold Change≥1.4), 4 vs. 0 Gy irradiation Genbank Fold Change P -Value Gene Symbol Description Accession Q9F8M7_CARHY (Q9F8M7) DTDP-glucose 4,6-dehydratase (Fragment), partial (9%) 6.70 0.017399678 THC2699065 [THC2719287] 5.53 0.003379195 BC013657 BC013657 Homo sapiens cDNA clone IMAGE:4152983, partial cds. [BC013657] 5.10 0.024641735 THC2750781 Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). 4.07 0.04353262 DNAH5 [Source:Uniprot/SWISSPROT;Acc:Q8TE73] [ENST00000382416] 3.81 0.002855909 NM_145263 SPATA18 Homo sapiens spermatogenesis associated 18 homolog (rat) (SPATA18), mRNA [NM_145263] AA418814 zw01a02.s1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:767978 3', 3.69 0.03203913 AA418814 AA418814 mRNA sequence [AA418814] AL356953 leucine-rich repeat-containing G protein-coupled receptor 6 {Homo sapiens} (exp=0; 3.63 0.0277936 THC2705989 wgp=1; cg=0), partial (4%) [THC2752981] AA484677 ne64a07.s1 NCI_CGAP_Alv1 Homo sapiens cDNA clone IMAGE:909012, mRNA 3.63 0.027098073 AA484677 AA484677 sequence [AA484677] oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence 3.48 0.04468495 AA837799 AA837799 [AA837799] Homo sapiens hypothetical protein LOC340109, mRNA (cDNA clone IMAGE:5578073), partial 3.27 0.031178378 BC039509 LOC643401 cds. [BC039509] Homo sapiens Fas (TNF receptor superfamily, member 6) (FAS), transcript variant 1, mRNA 3.24 0.022156298 NM_000043 FAS [NM_000043] 3.20 0.021043295 A_32_P125056 BF803942 CM2-CI0135-021100-477-g08 CI0135 Homo sapiens cDNA, mRNA sequence 3.04 0.043389246 BF803942 BF803942 [BF803942] 3.03 0.002430239 NM_015920 RPS27L Homo sapiens ribosomal protein S27-like (RPS27L), mRNA [NM_015920] Homo sapiens tumor necrosis factor receptor superfamily, member 10c, decoy without an 2.98 0.021202829 NM_003841 TNFRSF10C intracellular domain (TNFRSF10C), mRNA [NM_003841] 2.97 0.03243901 AB002384 C6orf32 Homo sapiens mRNA for KIAA0386 gene, partial cds. -

Pharmacological Targeting of the Mitochondrial Phosphatase PTPMT1 by Dahlia Doughty Shenton Department of Biochemistry Duke
Pharmacological Targeting of the Mitochondrial Phosphatase PTPMT1 by Dahlia Doughty Shenton Department of Biochemistry Duke University Date: May 1 st 2009 Approved: ___________________________ Dr. Patrick J. Casey, Supervisor ___________________________ Dr. Perry J. Blackshear ___________________________ Dr. Anthony R. Means ___________________________ Dr. Christopher B. Newgard ___________________________ Dr. John D. York Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Biochemistry in the Graduate School of Duke University 2009 ABSTRACT Pharmacological Targeting of the Mitochondrial Phosphatase PTPMT1 by Dahlia Doughty Shenton Department of Biochemistry Duke University Date: May 1 st 2009 Approved: ___________________________ Dr. Patrick J. Casey, Supervisor ___________________________ Dr. Perry J. Blackshear ___________________________ Dr. Anthony R. Means ___________________________ Dr. Christopher B. Newgard ___________________________ Dr. John D. York An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Biochemistry in the Graduate School of Duke University 2009 Copyright by Dahlia Doughty Shenton 2009 Abstract The dual specificity protein tyrosine phosphatases comprise the largest and most diverse group of protein tyrosine phosphatases and play integral roles in the regulation of cell signaling events. The dual specificity protein tyrosine phosphatases impact multiple -

Structural and Functional Analysis of PTPMT1, a Phosphatase Required for Cardiolipin Synthesis
Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis Junyu Xiaoa,1, James L. Engela,1, Ji Zhanga, Mark J. Chenb, Gerard Manningb, and Jack E. Dixona,c,d,e,2 aDepartment of Pharmacology, University of California, La Jolla, CA 92093; bRazavi Newman Center for Bioinformatics, Salk Institute, La Jolla, CA 92037; cDepartment of Cellular and Molecular Medicine, University of California, La Jolla, CA 92093; dDepartment of Chemistry and Biochemistry, University of California, La Jolla, CA 92093; and eHoward Hughes Medical Institute, Chevy Chase, MD 20815 Contributed by Jack E. Dixon, June 9, 2011 (sent for review December 29, 2010) PTPMT1 (PTP localized to the Mitochondrion 1) is a member of the chondria, and its level is not affected by the loss of PTPMT1, sug- protein tyrosine phosphatase superfamily that is localized exclu- gesting that PI(5)P is not its endogenous substrate (10). We sively to the mitochondrion. We recently reported that PTPMT1 recently demonstrated that the physiological target of PTPMT1 dephosphorylates phosphatidylglycerol phosphate, an essential is phosphatidylglycerol phosphate (PGP) (11), which is structu- intermediate of cardiolipin biosynthesis. To gain further insights rally remarkably similar to PI(5)P (Fig. 1C). into the molecular basis of PTPMT1 function, we determined the Phosphatidylglycerol (PG), the product of PTPMT1’s activity, crystal structures of the phosphatase domain of PTPMT1. PTPMT1 is an essential component of pulmonary surfactant, and a precur- exhibits a canonical protein tyrosine phosphatase domain fold, sor for cardiolipin biosynthesis (Fig. 1D and Fig. S2). Cardiolipin resembling many dual-specificity phosphatases such as phospha- is a glycerophospholipid found predominantly in the mitochon- tase and tensin homolog and vaccinia H1-related phosphatase. -

Revealing the Action Mechanisms of Dexamethasone on the Birth Weight
® Observational Study Medicine OPEN Revealing the action mechanisms of dexamethasone on the birth weight of infant using RNA-sequencing data of trophoblast cells ∗ Hongkai Shang, MDa, Liping Sun, BMa, Thorsten Braun, MDb, Qi Si, BMa, Jinyi Tong, MMa, Abstract Dexamethasone (DEX) could induce low birth weight of infant, and low birth weight has close associations with glucocorticoid levels, insulin resistance, hypertension, and metabolic syndrome in adulthood. This study was designed to reveal the action mechanisms of DEX on the birth weight of infant. Using quantitative real-time polymerase chain reaction (qRT-PCR), trophoblast cells of human placenta were identified and the optimum treatment time of DEX were determined. Trophoblast cells were treated by DEX (DEX group) or ethanol (control group) (each group had 3 samples), and then were performed with RNA-sequencing. Afterward, the differentially expressed genes (DEGs) were identified by R package, and their potential functions were successively enriched using DAVID database and Enrichr method. Followed by protein–protein interaction (PPI) network was constructed using Cytoscape software. Using Enrichr method and TargetScan software, the transcription factors (TFs) and micorRNAs (miRNAs) targeted the DEGs separately were predicted. Based on MsigDB database, gene set enrichment analysis (GSEA) was performed. There were 391 DEGs screened from the DEX group. Upregulated SRR and potassium voltage-gated channel subfamily J member 4(KCNJ4) and downregulated GALNT1 separately were enriched in PDZ (an acronym of PSD-95, Dlg, and ZO-1) domain binding and Mucin type O-glycan biosynthesis. In the PPI network, CDK2 and CDK4 had higher degrees. TFs ATF2 and E2F4 and miRNA miR-16 were predicted for the DEGs. -

Perspectives
FOCUS ON MECHANOTRANSDUCTION PERSPECTIVES network that can promote coordinated OPINION changes in cell, cytoskeletal and nuclear struc- ture in response to mechanical distortion14 Mechanotransduction at a (FIG. 1a). (Herein, the term hard-wired refers to cytoskeletal structures that are stable enough distance: mechanically coupling the as interconnected units to resist mechanical stresses and thereby maintain shape stabil- ity, even though they undergo continuous extracellular matrix with the nucleus dynamic remodelling at the molecular level.) This model takes into account the observa- Ning Wang, Jessica D. Tytell and Donald E. Ingber tion that individual cytoskeletal filaments can bear significant tensile and compressive loads Abstract | Research in cellular mechanotransduction often focuses on how in living cells because their structural integrity extracellular physical forces are converted into chemical signals at the cell surface. is maintained for longer than the turnover However, mechanical forces that are exerted on surface-adhesion receptors, such time of individual protein monomers15–17. as integrins and cadherins, are also channelled along cytoskeletal filaments and Key to the cellular tensegrity model is concentrated at distant sites in the cytoplasm and nucleus. Here, we explore the the idea that overall cell-shape stability and long-distance force transfer are governed by molecular mechanisms by which forces might act at a distance to induce the level of isometric tension, or ‘prestress’, mechanochemical conversion in the nucleus and alter gene activities. in the cytoskeleton that is generated through the establishment of a force balance between Mechanical forces influence the growth and For example, endothelial cells sense fluid opposing structural elements (that is, micro- shape of virtually every tissue and organ in shear through a cell–cell junctional com- tubules, contractile microfilaments and our bodies. -

Supplemental Table 1
Supplemental Data Supplemental Table 1. Genes differentially regulated by Ad-KLF2 vs. Ad-GFP infected EC. Three independent genome-wide transcriptional profiling experiments were performed, and significantly regulated genes were identified. Color-coding scheme: Up, p < 1e-15 Up, 1e-15 < p < 5e-10 Up, 5e-10 < p < 5e-5 Up, 5e-5 < p <.05 Down, p < 1e-15 As determined by Zpool Down, 1e-15 < p < 5e-10 Down, 5e-10 < p < 5e-5 Down, 5e-5 < p <.05 p<.05 as determined by Iterative Standard Deviation Algorithm as described in Supplemental Methods Ratio RefSeq Number Gene Name 1,058.52 KRT13 - keratin 13 565.72 NM_007117.1 TRH - thyrotropin-releasing hormone 244.04 NM_001878.2 CRABP2 - cellular retinoic acid binding protein 2 118.90 NM_013279.1 C11orf9 - chromosome 11 open reading frame 9 109.68 NM_000517.3 HBA2;HBA1 - hemoglobin, alpha 2;hemoglobin, alpha 1 102.04 NM_001823.3 CKB - creatine kinase, brain 96.23 LYNX1 95.53 NM_002514.2 NOV - nephroblastoma overexpressed gene 75.82 CeleraFN113625 FLJ45224;PTGDS - FLJ45224 protein;prostaglandin D2 synthase 21kDa 74.73 NM_000954.5 (brain) 68.53 NM_205545.1 UNQ430 - RGTR430 66.89 NM_005980.2 S100P - S100 calcium binding protein P 64.39 NM_153370.1 PI16 - protease inhibitor 16 58.24 NM_031918.1 KLF16 - Kruppel-like factor 16 46.45 NM_024409.1 NPPC - natriuretic peptide precursor C 45.48 NM_032470.2 TNXB - tenascin XB 34.92 NM_001264.2 CDSN - corneodesmosin 33.86 NM_017671.3 C20orf42 - chromosome 20 open reading frame 42 33.76 NM_024829.3 FLJ22662 - hypothetical protein FLJ22662 32.10 NM_003283.3 TNNT1 - troponin T1, skeletal, slow LOC388888 (LOC388888), mRNA according to UniGene - potential 31.45 AK095686.1 CONFLICT - LOC388888 (na) according to LocusLink. -

Network of Micrornas-Mrnas Interactions in Pancreatic Cancer
Hindawi Publishing Corporation BioMed Research International Volume 2014, Article ID 534821, 8 pages http://dx.doi.org/10.1155/2014/534821 Research Article Network of microRNAs-mRNAs Interactions in Pancreatic Cancer Elnaz Naderi,1 Mehdi Mostafaei,2 Akram Pourshams,1 and Ashraf Mohamadkhani1 1 Liver and Pancreatobiliary Diseases Research Center, Digestive Diseases Research Institute, Tehran University of Medical Sciences, Tehran, Iran 2 Biotechnology Engineering, Islamic Azad University,Tehran North Branch, Tehran, Iran Correspondence should be addressed to Ashraf Mohamadkhani; [email protected] Received 5 February 2014; Revised 13 April 2014; Accepted 13 April 2014; Published 7 May 2014 Academic Editor: FangXiang Wu Copyright © 2014 Elnaz Naderi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. MicroRNAs are small RNA molecules that regulate the expression of certain genes through interaction with mRNA targets and are mainly involved in human cancer. This study was conducted to make the network of miRNAs-mRNAs interactions in pancreatic cancer as the fourth leading cause of cancer death. Methods. 56 miRNAs that were exclusively expressed and 1176 genes that were downregulated or silenced in pancreas cancer were extracted from beforehand investigations. MiRNA–mRNA interactions data analysis and related networks were explored using MAGIA tool and Cytoscape 3 software. Functional annotations of candidate genes in pancreatic cancer were identified by DAVID annotation tool. Results. This network is made of 217 nodes for mRNA, 15 nodes for miRNA, and 241 edges that show 241 regulations between 15 miRNAs and 217 target genes. -

The SUN Rises on Meiotic Chromosome Dynamics
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Developmental Cell Review Feature The SUN Rises on Meiotic Chromosome Dynamics Yasushi Hiraoka1,2,* and Abby F. Dernburg3,4,* 1Graduate School of Frontier Biosciences, Osaka University, 1-3 Yamadaoka, Suita 565-0871, Japan 2Kobe Advanced ICT Research Center, National Institute of Information and Communications Technology, 588-2 Iwaoka, Iwaoka-cho, Nishi-ku, Kobe 651-2492, Japan 3Howard Hughes Medical Institute and Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA 94720-3220, USA 4Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA *Correspondence: [email protected] (Y.H.), [email protected] (A.F.D.) DOI 10.1016/j.devcel.2009.10.014 Recent studies in diverse eukaryotes have implicated a family of nuclear envelope proteins containing SUN domains as key components of meiotic nuclear organization and chromosome dynamics. In many cases, these transmembrane proteins are also known to contribute to centrosome or spindle pole body function in mitotically dividing cells. During meiotic prophase, the apparent role of these SUN-domain proteins, together with their partner KASH-domain proteins, is to connect chromosomes through the intact nuclear envelope to force-generating mechanisms in the cytoplasm. A Family of SUN Proteins In this review, we use these existing names and also refer to these The SUN domain was named for homologous sequences shared genes and their orthologs in mouse and human genomes as Sun4 by Sad1 and UNC-84 proteins (Malone et al., 1999). -
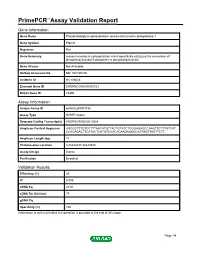
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name Phosphatidylglycerophosphatase and protein-tyrosine phosphatase 1 Gene Symbol Ptpmt1 Organism Rat Gene Summary mouse homolog is a phosphatase which specifically catalyzes the conversion of phosphatidylinositol 5-phosphate to phosphatidylinositol Gene Aliases Not Available RefSeq Accession No. NM_001105726 UniGene ID Rn.108023 Ensembl Gene ID ENSRNOG00000009723 Entrez Gene ID 29390 Assay Information Unique Assay ID qRnoCED0003185 Assay Type SYBR® Green Detected Coding Transcript(s) ENSRNOT00000013584 Amplicon Context Sequence AGCGCTTTGTCCTTTAACATGTTACTGTATCTCGGAGAGCCAAGTTCTTTGTCAT CCACAGACTTCATACTCATGTCAGCACAAGAGACCAGTAGTTAGTTCTT Amplicon Length (bp) 74 Chromosome Location 3:86444437-86444540 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 99 R2 0.999 cDNA Cq 20.91 cDNA Tm (Celsius) 79 gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Ptpmt1, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Identification of Potential Core Genes in Sevoflurance Induced Myocardial
Identication of Potential core genes in Sevourance induced Myocardial Energy Metabolism in Patients Undergoing Off-pump Coronary Artery Bypass Graft Surgery using Bioinformatics analysis Hua Lin ( [email protected] ) Tianjin Medical University General Hospital Airport Site Research article Keywords: sevourane, Myocardial Energy Metabolism, Off-pump Coronary Artery Bypass Graft Surgery Posted Date: November 18th, 2019 DOI: https://doi.org/10.21203/rs.2.17434/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/15 Abstract Background: Myocardial ischemia-reperfusion injury always happened after Off-pump coronary artery bypass graft(OPCABG), and this can not be avoided altogether. In this study, we tried to detect potential genes of sevourane-induced myocardial energy metabolism in patients undergoing OPCABG using bioinformatics analysis. Methods: We download and analyze the gene expression prole data from the Gene Expression Omnibus(GEO) database using bioinformatics methods. We downloded the gene expression data from the Gene Expression Omnibus(GEO) database using bioinformatics methods. Gene Ontology(GO) functional annotation analysis and Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway enrichment analysis were used to analysis the screened differentially expressed genes(DEGs). Then, we established a protein–protein interaction (PPI) network to nd hub genes associated with myocardial energy metabolism. Results: Through PPI network, we nd ten hub genes, including JUN, EGR1, ATF3, FOSB, JUNB, DUSP1, EGR2, NR4A1, BTG2, NR4A2. Conclusions: In conclusion, the proteins encoded by EGR1ATF3c-FosBtg2JunBDUSP1NR4A1BTG2 and NR4A2 were related to cardiac function. ATF3, FOSB, JUNB, DUSP1, NR4A1, NR4A2 are related to apoptosis of cardiomyocytes. The protein encoded by BTG2 is related to hypertrophy.