Proceedings of the 11Th Python in Science Conference
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Graham Holdings Company 2014 Annual Report
GRAHAM HOLDINGS 2014 ANNUAL REPORT REVENUE BY PRINCIPAL OPERATIONS n EDUCATION 61% n CABLE 23% n TELEVISION BROADCASTING 10% n OTHER BUSINESSES 6% FINANCIAL HIGHLIGHTS (in thousands, except per share amounts) 2014 2013 Change Operating revenues $ 3,535,166 $ 3,407,911 4% Income from operations $ 407,932 $ 319,169 28% Net income attributable to common shares $ 1,292,996 $ 236,010 — Diluted earnings per common share from continuing operations $ 138.88 $ 23.36 — Diluted earnings per common share $ 195.03 $ 32.05 — Dividends per common share $ 10.20 $ — — Common stockholders’ equity per share $ 541.54 $ 446.73 21% Diluted average number of common shares outstanding 6,559 7,333 –11% INCOME FROM NET INCOME ATTRIBUTABLE OPERATING REVENUES OPERATIONS TO COMMON SHARES ($ in millions) ($ in millions) ($ in millions) 3,861 582 1,293 3,453 3,535 3,373 3,408 408 314 319 149 277 236 116 131 2010 2011 2012 2013 2014 2010 2011 2012 2013 2014 2010 2011 2012 2013 2014 RETURN ON DILUTED EARNINGS PER AVERAGE COMMON COMMON SHARE FROM DILUTED EARNINGS STOCKHOLDERS’ EQUITY* CONTINUING OPERATIONS PER COMMON SHARE ($) ($) 46.6% 138.88 195.03 38.16 9.8% 9.0% 23.36 31.04 32.05 5.2% 17.32 4.4% 14.70 17.39 6.40 2010 2011 2012 2013 2014 2010 2011 2012 2013 2014 2010 2011 2012 2013 2014 * Computed on a comparable basis, excluding the impact of the adjustment for pensions and other postretirement plans on average common stockholders’ equity. 2014 ANNUAL REPORT 1 To OUR SHAREHOLDERS Quite a lot happened in 2014. -

Can We Rule out the Possibility of the Existence of an Additional Time Axis
Brian D. Koberlein Thomas J. Murray Time 2 ABSTRACT CAN WE RULE OUT THE POSSIBILITY THAT THERE IS MORE THAN ONE DIRECTION IN TIME? by Lynn Stephens The literature was examined to see if a proposal for the existence of additional time-like axes could be falsified. Some naïve large-time models are visualized and examined from the standpoint of simplicity and adequacy. Although the conclusion here is that the existence of additional time-like dimensions has not been ruled out, the idea is probably not falsifiable, as was first assumed. The possibility of multiple times has been dropped from active consideration by theorists less because of negative evidence than because it can lead to cumbersome models that introduce causality problems without providing any advantages over other models. Even though modern philosophers have made intriguing suggestions concerning the causality issue, usability and usefulness issues remain. Symmetry considerations from special relativity and from the conservation laws are used to identify restrictions on large-time models. In view of recent experimental findings, further research into Cramer’s transactional model is indicated. This document contains embedded graphic (.png and .jpg) and Apple Quick Time (.mov) files. The CD-ROM contains interactive Pacific Tech Graphing Calculator (.gcf) files. Time 3 NOTE TO THE READER The graphics in the PDF version of this document rely heavily on the use of color and will not print well in black and white. The CD-ROM attachment that comes with the paper version of this document includes: • The full-color PDF with QuickTime animation; • A PDF printable in black and white; • The original interactive Graphing Calculator files. -

Martha L. Minow
Martha L. Minow 1525 Massachusetts Avenue Griswold 407, Harvard Law School Cambridge, MA 02138 (617) 495-4276 [email protected] Current Academic Appointments: 300th Anniversary University Professor, Harvard University Harvard University Distinguished Service Professor Faculty, Harvard Graduate School of Education Faculty Associate, Carr Center for Human Rights, Harvard Kennedy School of Government Current Activities: Advantage Testing Foundation, Vice-Chair and Trustee American Academy of Arts and Sciences, Access to Justice Project American Bar Association Center for Innovation, Advisory Council American Law Institute, Member Berkman Klein Center for Internet and Society, Harvard University, Director Campaign Legal Center, Board of Trustees Carnegie Corporation, Board of Trustees Committee to Visit the Harvard Business School, Harvard University Board of Overseers Facing History and Ourselves, Board of Scholars Harvard Data Science Review, Associate Editor Initiative on Harvard and the Legacy of Slavery Law, Violence, and Meaning Series, Univ. of Michigan Press, Co-Editor MacArthur Foundation, Director MIT Media Lab, Advisory Council MIT Schwarzman College of Computing, Co-Chair, External Advisory Council National Academy of Sciences' Committee on Science, Technology, and Law Profiles in Courage Award Selection Committee, JFK Library, Chair Russell Sage Foundation, Trustee Skadden Fellowship Foundation, Selection Trustee Susan Crown Exchange Foundation, Trustee WGBH Board of Trustees, Trustee Education: Yale Law School, J.D. 1979 Articles and Book Review Editor, Yale Law Journal, 1978-1979 Editor, Yale Law Journal, 1977-1978 Harvard Graduate School of Education, Ed.M. 1976 University of Michigan, A.B. 1975 Phi Beta Kappa, Magna Cum Laude James B. Angell Scholar, Branstrom Prize New Trier East High School, Winnetka, Illinois, 1968-1972 Honors and Fellowships: Leo Baeck Medal, Nov. -

Rutherford's Nuclear World: the Story of the Discovery of the Nuc
Rutherford's Nuclear World: The Story of the Discovery of the Nuc... http://www.aip.org/history/exhibits/rutherford/sections/atop-physic... HOME SECTIONS CREDITS EXHIBIT HALL ABOUT US rutherford's explore the atom learn more more history of learn about aip's nuclear world with rutherford about this site physics exhibits history programs Atop the Physics Wave ShareShareShareShareShareMore 9 RUTHERFORD BACK IN CAMBRIDGE, 1919–1937 Sections ← Prev 1 2 3 4 5 Next → In 1962, John Cockcroft (1897–1967) reflected back on the “Miraculous Year” ( Annus mirabilis ) of 1932 in the Cavendish Laboratory: “One month it was the neutron, another month the transmutation of the light elements; in another the creation of radiation of matter in the form of pairs of positive and negative electrons was made visible to us by Professor Blackett's cloud chamber, with its tracks curled some to the left and some to the right by powerful magnetic fields.” Rutherford reigned over the Cavendish Lab from 1919 until his death in 1937. The Cavendish Lab in the 1920s and 30s is often cited as the beginning of modern “big science.” Dozens of researchers worked in teams on interrelated problems. Yet much of the work there used simple, inexpensive devices — the sort of thing Rutherford is famous for. And the lab had many competitors: in Paris, Berlin, and even in the U.S. Rutherford became Cavendish Professor and director of the Cavendish Laboratory in 1919, following the It is tempting to simplify a complicated story. Rutherford directed the Cavendish Lab footsteps of J.J. Thomson. Rutherford died in 1937, having led a first wave of discovery of the atom. -

Jul/Aug 2013
I NTERNATIONAL J OURNAL OF H IGH -E NERGY P HYSICS CERNCOURIER WELCOME V OLUME 5 3 N UMBER 6 J ULY /A UGUST 2 0 1 3 CERN Courier – digital edition Welcome to the digital edition of the July/August 2013 issue of CERN Courier. This “double issue” provides plenty to read during what is for many people the holiday season. The feature articles illustrate well the breadth of modern IceCube brings particle physics – from the Standard Model, which is still being tested in the analysis of data from Fermilab’s Tevatron, to the tantalizing hints of news from the deep extraterrestrial neutrinos from the IceCube Observatory at the South Pole. A connection of a different kind between space and particle physics emerges in the interview with the astronaut who started his postgraduate life at CERN, while connections between particle physics and everyday life come into focus in the application of particle detectors to the diagnosis of breast cancer. And if this is not enough, take a look at Summer Bookshelf, with its selection of suggestions for more relaxed reading. To sign up to the new issue alert, please visit: http://cerncourier.com/cws/sign-up. To subscribe to the magazine, the e-mail new-issue alert, please visit: http://cerncourier.com/cws/how-to-subscribe. ISOLDE OUTREACH TEVATRON From new magic LHC tourist trail to the rarest of gets off to a LEGACY EDITOR: CHRISTINE SUTTON, CERN elements great start Results continue DIGITAL EDITION CREATED BY JESSE KARJALAINEN/IOP PUBLISHING, UK p6 p43 to excite p17 CERNCOURIER www. -

Simulating Physics with Computers
International Journal of Theoretical Physics, VoL 21, Nos. 6/7, 1982 Simulating Physics with Computers Richard P. Feynman Department of Physics, California Institute of Technology, Pasadena, California 91107 Received May 7, 1981 1. INTRODUCTION On the program it says this is a keynote speech--and I don't know what a keynote speech is. I do not intend in any way to suggest what should be in this meeting as a keynote of the subjects or anything like that. I have my own things to say and to talk about and there's no implication that anybody needs to talk about the same thing or anything like it. So what I want to talk about is what Mike Dertouzos suggested that nobody would talk about. I want to talk about the problem of simulating physics with computers and I mean that in a specific way which I am going to explain. The reason for doing this is something that I learned about from Ed Fredkin, and my entire interest in the subject has been inspired by him. It has to do with learning something about the possibilities of computers, and also something about possibilities in physics. If we suppose that we know all the physical laws perfectly, of course we don't have to pay any attention to computers. It's interesting anyway to entertain oneself with the idea that we've got something to learn about physical laws; and if I take a relaxed view here (after all I'm here and not at home) I'll admit that we don't understand everything. -

A Selected Bibliography of Publications By, and About, J
A Selected Bibliography of Publications by, and about, J. Robert Oppenheimer Nelson H. F. Beebe University of Utah Department of Mathematics, 110 LCB 155 S 1400 E RM 233 Salt Lake City, UT 84112-0090 USA Tel: +1 801 581 5254 FAX: +1 801 581 4148 E-mail: [email protected], [email protected], [email protected] (Internet) WWW URL: http://www.math.utah.edu/~beebe/ 17 March 2021 Version 1.47 Title word cross-reference $1 [Duf46]. $12.95 [Edg91]. $13.50 [Tho03]. $14.00 [Hug07]. $15.95 [Hen81]. $16.00 [RS06]. $16.95 [RS06]. $17.50 [Hen81]. $2.50 [Opp28g]. $20.00 [Hen81, Jor80]. $24.95 [Fra01]. $25.00 [Ger06]. $26.95 [Wol05]. $27.95 [Ger06]. $29.95 [Goo09]. $30.00 [Kev03, Kle07]. $32.50 [Edg91]. $35 [Wol05]. $35.00 [Bed06]. $37.50 [Hug09, Pol07, Dys13]. $39.50 [Edg91]. $39.95 [Bad95]. $8.95 [Edg91]. α [Opp27a, Rut27]. γ [LO34]. -particles [Opp27a]. -rays [Rut27]. -Teilchen [Opp27a]. 0-226-79845-3 [Guy07, Hug09]. 0-8014-8661-0 [Tho03]. 0-8047-1713-3 [Edg91]. 0-8047-1714-1 [Edg91]. 0-8047-1721-4 [Edg91]. 0-8047-1722-2 [Edg91]. 0-9672617-3-2 [Bro06, Hug07]. 1 [Opp57f]. 109 [Con05, Mur05, Nas07, Sap05a, Wol05, Kru07]. 112 [FW07]. 1 2 14.99/$25.00 [Ber04a]. 16 [GHK+96]. 1890-1960 [McG02]. 1911 [Meh75]. 1945 [GHK+96, Gow81, Haw61, Bad95, Gol95a, Hew66, She82, HBP94]. 1945-47 [Hew66]. 1950 [Ano50]. 1954 [Ano01b, GM54, SZC54]. 1960s [Sch08a]. 1963 [Kuh63]. 1967 [Bet67a, Bet97, Pun67, RB67]. 1976 [Sag79a, Sag79b]. 1981 [Ano81]. 20 [Goe88]. 2005 [Dre07]. 20th [Opp65a, Anoxx, Kai02]. -

Luis Alvarez: the Ideas Man
CERN Courier March 2012 Commemoration Luis Alvarez: the ideas man The years from the early 1950s to the late 1980s came alive again during a symposium to commemorate the birth of one of the great scientists and inventors of the 20th century. Luis Alvarez – one of the greatest experimental physicists of the 20th century – combined the interests of a scientist, an inventor, a detective and an explorer. He left his mark on areas that ranged from radar through to cosmic rays, nuclear physics, particle accel- erators, detectors and large-scale data analysis, as well as particles and astrophysics. On 19 November, some 200 people gathered at Berkeley to commemorate the 100th anniversary of his birth. Alumni of the Alvarez group – among them physicists, engineers, programmers and bubble-chamber film scanners – were joined by his collaborators, family, present-day students and admirers, as well as scientists whose professional lineage traces back to him. Hosted by the Lawrence Berkeley National Laboratory (LBNL) and the University of California at Berkeley, the symposium reviewed his long career and lasting legacy. A recurring theme of the symposium was, as one speaker put it, a “Shakespeare-type dilemma”: how could one person have accom- plished all of that in one lifetime? Beyond his own initiatives, Alvarez created a culture around him that inspired others to, as George Smoot put it, “think big,” as well as to “think broadly and then deep” and to take risks. Combined with Alvarez’s strong scientific standards and great care in execut- ing them, these principles led directly to the awarding of two Nobel Luis Alvarez celebrating the announcement of his 1968 Nobel prizes in physics to scientists at Berkeley – George Smoot in 2006 prize. -

1 / 3 Anwendungen Von Festkörperphysik 1 / 4
1 / 3 Anwendungen von Festkörperphysik Elektronik/Optoelektronik 1 / 4 Anwendungen von Festkörperphysik Organische (Opto-)elektronik Selbstreinigende Oberflächen Energietechnologie 2 1 / 5 Anwendungen von Festkörperphysik „intelligente“ Materialien Schaltbare Molekülschichten Magnetoelektrische Sensoren „Brain-Maschine-Interface“ 1 / 6 Nobelpreise für Physik zu festkörperphysikalischen Themen 1913 Heike Kamerlingh Onnes 1914 Max von Laue 1915 William Henry Bragg, William Lawrence Bragg 1920 Charles Edouard Guillaume 1921 Albert Einstein 1923 Robert Andrews Millikan Details siehe 1924 Karl Manne Siegbahn 1926 Jean Baptiste Perrin http://almaz.com/nobel/ 1937 Clinton Davisson, George Paget physics/physics.html 1946 Percy W. Bridgman 1956 William B. Shockley, John Bardeen und Walter H. Brattain 1961 Rudolf Mößbauer 1962 Lev Landau 1971 Louis Néel 1972 John Bardeen, Leon Neil Cooper, Robert Schrieffer 1973 Leo Esaki, Ivar Giaever, Brian Davon Josephson 1977 Philip W. Anderson, Nevill F. Mott, John H. van Vleck 1978 Pjotr Kapiza 1982 Kenneth G. Wilson 1985 Klaus von Klitzing 1986 Ernst Ruska, Gerd Binnig, Heinrich Rohrer 1987 Johannes Georg Bednorz, Karl Alex Müller 1991 Pierre-Gilles de Gennes 1994 Bertram N. Brockhouse, Clifford Glenwood Shull 1996 David M. Lee, Douglas D. Osheroff, Robert C. Richardson 1998 Robert B. Laughlin, Horst Ludwig Störmer, Daniel Chee Tsui 2000 Schores Alfjorow, Herbert Kroemer, Jack S. Kilby 2001 Eric A. Cornell, Wolfgang Ketterle, Carl E. Wieman 2003 Alexei Abrikossow, Witali Ginsburg, Anthony James Leggett 2007 -

A Suburb Unlike Any Other: a History of Early Jewish Life in Los Alamos, New Mexico
A Suburb Unlike Any Other: A History of Early Jewish Life in Los Alamos, New Mexico Master’s Thesis Presented to The Faculty of the Graduate School of Arts and Sciences Brandeis University Department of Near Eastern and Judaic Studies Jonathan Krasner, Jonathan Sarna, Advisors In Partial Fulfillment of the Requirements for the Degree Master of Arts in Near Eastern and Judaic Studies by Gabriel Weinstein May 2018 Copyright by Gabriel Weinstein © 2018 ABSTRACT A Suburb Unlike Any Other: A History of Early Jewish Life in Los Alamos, New Mexico A thesis presented to the Department of Near Eastern and Judaic Studies Graduate School of Arts and Sciences Brandeis University Waltham, Massachusetts By Gabriel Weinstein This thesis examines the development of the Jewish community in Los Alamos, New Mexico during the 1940s and 1950s and compares its development with Jewish communities located near federally funded scientific research centers. The Jewish community of Los Alamos formed during World War II as over one hundred scientists from around the world arrived to work on the development of the atomic bomb for the Manhattan Project. When the war ended, dozens of Jewish scientists remained at the Los Alamos Scientific Laboratory and established a permanent Jewish community. As the federal government transformed Los Alamos into a suburb in the 1950s, its Jewish community adapted many elements of suburban Jewish life. This thesis examines how Los Alamos Jews integrated socially and culturally into Los Alamos’ diverse cultural milieu and how Jewish -
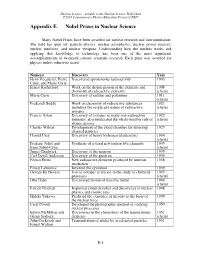
Appendix E Nobel Prizes in Nuclear Science
Nuclear Science—A Guide to the Nuclear Science Wall Chart ©2018 Contemporary Physics Education Project (CPEP) Appendix E Nobel Prizes in Nuclear Science Many Nobel Prizes have been awarded for nuclear research and instrumentation. The field has spun off: particle physics, nuclear astrophysics, nuclear power reactors, nuclear medicine, and nuclear weapons. Understanding how the nucleus works and applying that knowledge to technology has been one of the most significant accomplishments of twentieth century scientific research. Each prize was awarded for physics unless otherwise noted. Name(s) Discovery Year Henri Becquerel, Pierre Discovered spontaneous radioactivity 1903 Curie, and Marie Curie Ernest Rutherford Work on the disintegration of the elements and 1908 chemistry of radioactive elements (chem) Marie Curie Discovery of radium and polonium 1911 (chem) Frederick Soddy Work on chemistry of radioactive substances 1921 including the origin and nature of radioactive (chem) isotopes Francis Aston Discovery of isotopes in many non-radioactive 1922 elements, also enunciated the whole-number rule of (chem) atomic masses Charles Wilson Development of the cloud chamber for detecting 1927 charged particles Harold Urey Discovery of heavy hydrogen (deuterium) 1934 (chem) Frederic Joliot and Synthesis of several new radioactive elements 1935 Irene Joliot-Curie (chem) James Chadwick Discovery of the neutron 1935 Carl David Anderson Discovery of the positron 1936 Enrico Fermi New radioactive elements produced by neutron 1938 irradiation Ernest Lawrence -

An Improbable Venture
AN IMPROBABLE VENTURE A HISTORY OF THE UNIVERSITY OF CALIFORNIA, SAN DIEGO NANCY SCOTT ANDERSON THE UCSD PRESS LA JOLLA, CALIFORNIA © 1993 by The Regents of the University of California and Nancy Scott Anderson All rights reserved. Library of Congress Cataloging in Publication Data Anderson, Nancy Scott. An improbable venture: a history of the University of California, San Diego/ Nancy Scott Anderson 302 p. (not including index) Includes bibliographical references (p. 263-302) and index 1. University of California, San Diego—History. 2. Universities and colleges—California—San Diego. I. University of California, San Diego LD781.S2A65 1993 93-61345 Text typeset in 10/14 pt. Goudy by Prepress Services, University of California, San Diego. Printed and bound by Graphics and Reproduction Services, University of California, San Diego. Cover designed by the Publications Office of University Communications, University of California, San Diego. CONTENTS Foreword.................................................................................................................i Preface.........................................................................................................................v Introduction: The Model and Its Mechanism ............................................................... 1 Chapter One: Ocean Origins ...................................................................................... 15 Chapter Two: A Cathedral on a Bluff ......................................................................... 37 Chapter Three: