Computational Tutorials with Cantera
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Package Name Software Description Project
A S T 1 Package Name Software Description Project URL 2 Autoconf An extensible package of M4 macros that produce shell scripts to automatically configure software source code packages https://www.gnu.org/software/autoconf/ 3 Automake www.gnu.org/software/automake 4 Libtool www.gnu.org/software/libtool 5 bamtools BamTools: a C++ API for reading/writing BAM files. https://github.com/pezmaster31/bamtools 6 Biopython (Python module) Biopython is a set of freely available tools for biological computation written in Python by an international team of developers www.biopython.org/ 7 blas The BLAS (Basic Linear Algebra Subprograms) are routines that provide standard building blocks for performing basic vector and matrix operations. http://www.netlib.org/blas/ 8 boost Boost provides free peer-reviewed portable C++ source libraries. http://www.boost.org 9 CMake Cross-platform, open-source build system. CMake is a family of tools designed to build, test and package software http://www.cmake.org/ 10 Cython (Python module) The Cython compiler for writing C extensions for the Python language https://www.python.org/ 11 Doxygen http://www.doxygen.org/ FFmpeg is the leading multimedia framework, able to decode, encode, transcode, mux, demux, stream, filter and play pretty much anything that humans and machines have created. It supports the most obscure ancient formats up to the cutting edge. No matter if they were designed by some standards 12 ffmpeg committee, the community or a corporation. https://www.ffmpeg.org FFTW is a C subroutine library for computing the discrete Fourier transform (DFT) in one or more dimensions, of arbitrary input size, and of both real and 13 fftw complex data (as well as of even/odd data, i.e. -

RMG-Py and Cantherm Documentation ⇌Release 2.0.0
RMG RMG-Py and CanTherm Documentation ⇌Release 2.0.0 William H. Green, Richard H. West, and the RMG Team Sep 16, 2016 CONTENTS 1 RMG User’s Guide 3 1.1 Introduction...............................................3 1.2 Release Notes..............................................4 1.3 Overview of Features...........................................6 1.4 Installation................................................7 1.5 Creating Input Files........................................... 20 1.6 Example Input Files........................................... 29 1.7 Running a Job.............................................. 36 1.8 Analyzing the Output Files........................................ 37 1.9 Species Representation.......................................... 38 1.10 Group Representation.......................................... 39 1.11 Databases................................................. 39 1.12 Thermochemistry Estimation...................................... 58 1.13 Kinetics Estimation........................................... 63 1.14 Liquid Phase Systems.......................................... 65 1.15 Guidelines for Building a Model..................................... 72 1.16 Standalone Modules........................................... 73 1.17 Frequently Asked Questions....................................... 81 1.18 Credits.................................................. 82 1.19 How to Cite................................................ 82 2 CanTherm User’s Guide 83 2.1 Introduction.............................................. -

Pyjac: Analytical Jacobian Generator for Chemical Kinetics
pyJac: analytical Jacobian generator for chemical kinetics Kyle E. Niemeyera,∗, Nicholas J. Curtisb, Chih-Jen Sungb aSchool of Mechanical, Industrial, and Manufacturing Engineering Oregon State University, Corvallis, OR 97331, USA bDepartment of Mechanical Engineering University of Connecticut, Storrs, CT, 06269, USA Abstract Accurate simulations of combustion phenomena require the use of detailed chemical kinet- ics in order to capture limit phenomena such as ignition and extinction as well as predict pollutant formation. However, the chemical kinetic models for hydrocarbon fuels of prac- tical interest typically have large numbers of species and reactions and exhibit high levels of mathematical stiffness in the governing differential equations, particularly for larger fuel molecules. In order to integrate the stiff equations governing chemical kinetics, generally reactive-flow simulations rely on implicit algorithms that require frequent Jacobian matrix evaluations. Some in situ and a posteriori computational diagnostics methods also require accurate Jacobian matrices, including computational singular perturbation and chemical ex- plosive mode analysis. Typically, finite differences numerically approximate these, but for larger chemical kinetic models this poses significant computational demands since the num- ber of chemical source term evaluations scales with the square of species count. Furthermore, existing analytical Jacobian tools do not optimize evaluations or support emerging SIMD processors such as GPUs. Here we introduce pyJac, a Python-based open-source program that generates analytical Jacobian matrices for use in chemical kinetics modeling and anal- ysis. In addition to producing the necessary customized source code for evaluating reaction rates (including all modern reaction rate formulations), the chemical source terms, and the Jacobian matrix, pyJac uses an optimized evaluation order to minimize computational and memory operations. -
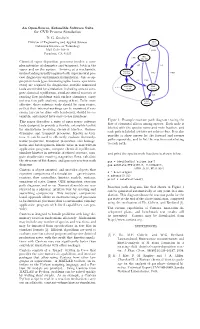
An Open-Source, Extensible Software Suite for CVD Process Simulation D
An Open-Source, Extensible Software Suite for CVD Process Simulation D. G. Goodwin Division of Engineering and Applied Science SIH4 0.0016 California Institute of Technology Mail Code 104-44 0.0135 Pasadena, CA 91125 SIH2 0.852 4.8e-005 Chemical vapor deposition processes involve a com- plex interplay of chemistry and transport, both in the 0.85 0.4 vapor and on the surface. Arriving at a mechanistic H3SISIH 0.415 0.429 understanding usually requires both experimental pro- 2.9e-005 cess diagnostics and numerical simulation. Just as ap- propriate tools (gas chromatographs, lasers, spectrom- 1 0.8 0.429 eters) are required for diagnostics, suitable numerical H2SISIH2 SI3H8 0.101 tools are needed for simulation, including ones to com- 0.0038 7.3e-005 pute chemical equilibrium, simulate stirred reactors or reacting flow problems with surface chemistry, carry 0.83 out reaction path analysis, among others. To be most SI2H6 effective, these software tools should be open-source, 0.00019 so that their internal workings can be examined if nec- essary (as can be done with hardware), should be ex- tensible, and should have easy-to-use interfaces. Figure 1: Example reaction path diagram tracing the This paper describes a suite of open-source software flow of elemental silicon among species. Each node is tools designed to provide a flexible, extensible toolkit labeled with the species name and mole fraction, and for simulations involving chemical kinetics, thermo- each path is labeled with its net relative flux. It is also dynamics, and transport processes. Known as Can- possible to show arrows for the forward and reverse tera, it can be used to efficiently evaluate thermody- paths separately, and to list the reaction contributing namic properties, transport properties, and homoge- to each path. -

BIOVIA MATERIALS STUDIO OVERVIEW Datasheet
BIOVIA MATERIALS STUDIO OVERVIEW Datasheet 10 Å 3 nm 15 µm Modeling and Simulation for Next Generation Materials. BIOVIA Materials Studio® is a complete modeling and simulation environment that enables researchers in materials science and chemistry to develop new materials by predicting the relationships of a material’s atomic and molecular structure with its properties and behavior. Using Materials Studio, researchers in many industries can engineer better performing materials of all types, including pharmaceuticals, catalysts, polymers and composites, metals and alloys, batteries and fuel cells, nanomaterials, and more. Materials Studio is the world’s most advanced, yet easy Materials Visualizer provides capability to construct, manipulate to use environment for modeling and evaluating materials and view models of molecules, crystalline materials, surfaces, performance and behavior. Using Materials Studio, materials polymers, and mesoscale structures. It also supports the full scientists experience the following benefits: range of Materials Studio simulations with capabilities to • Reduction in cost and time associated with physical testing visualize results through images, animations, graphs, charts, and experimentation through “Virtual Screening” of candidate tables, and textual data. Most tools in the Materials Visualizer material variations. can also be accessed through the MaterialsScript API, allowing • Acceleration of the innovation process - developing new, better expert users to create custom capabilities and automate performing, more sustainable, and cost effective materials faster repetitive tasks. Materials Visualizer’s Microsoft Windows than can be done with physical testing and experimentation. client operates with a range of Windows and Linux server • Improved fundamental understanding of the relationship architectures to provide a highly responsive user experience. between atomic and molecular structure with material properties and behavior. -

Quick Navigation Mechanical Engineering Department Facilities
Quick Navigation Department Facilities Multi-User Facilities Other Facilities Mechanical Engineering Department Facilities Departmental Facilities and Individual Faculty Research Laboratories: Shared Services Facilities Director: Haejune Kim We provide students and researchers open access to a wide range of equipment for measuring, characterizing and imaging of samples. If necessary, shared facility staff or super user of the instrument will offer training before giving access to the equipment. The shared services facilities are available to mechanical engineering faculty and students. Currently the group maintains and has available for scheduling the following machines:VEGA II LSU SEM, KLA- Tencor P-6 Stylus Profiler, VHX-600 Digital Microscope, High Performance Liquid Chromatography (HPLC), Total Organic Carbon (TOC) Analyzer, Atomic Layer Deposition (ALD) System, MicroClimate environmental chamber, Photron FSTCAM SA5 High Speed Camera, Olympus BX 61/Visitech QLC-100 Confocal Microscopy, Simultaneous Thermal Analyzer – Q600 SDT, Differential Scanning Calorimeter – DSC Q1000, Veeco Dektak 150 Profilometer, and Salt Spray Chamber. Acoustics and Signal Processing Laboratory Contact: Dr. Yong-Joe Kim The ASPL, founded in 2009, is a research laboratory of the Department of Mechanical Engineering at Texas A&M University (TAMU), College Station, Texas, USA. It is located at James J. Cain ’51 Building #409. Prof. Yong-Joe Kim currently serves as the director for the ASPL. The current research is focused on the areas of acoustics, signal processing, vibration, dynamics, and biomechanics. Advanced Computational Mechanics Laboratory Contact: Dr. J.N. Reddy Professor Reddy’s Advanced Computational Mechanics Laboratory (ACML) at Texas A&M University is dedicated to state-of-the-art research in the development of novel mathematical models and numerical simulation of physical phenomena. -

A Partial Glossary of Spanish Geological Terms Exclusive of Most Cognates
U.S. DEPARTMENT OF THE INTERIOR U.S. GEOLOGICAL SURVEY A Partial Glossary of Spanish Geological Terms Exclusive of Most Cognates by Keith R. Long Open-File Report 91-0579 This report is preliminary and has not been reviewed for conformity with U.S. Geological Survey editorial standards or with the North American Stratigraphic Code. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. 1991 Preface In recent years, almost all countries in Latin America have adopted democratic political systems and liberal economic policies. The resulting favorable investment climate has spurred a new wave of North American investment in Latin American mineral resources and has improved cooperation between geoscience organizations on both continents. The U.S. Geological Survey (USGS) has responded to the new situation through cooperative mineral resource investigations with a number of countries in Latin America. These activities are now being coordinated by the USGS's Center for Inter-American Mineral Resource Investigations (CIMRI), recently established in Tucson, Arizona. In the course of CIMRI's work, we have found a need for a compilation of Spanish geological and mining terminology that goes beyond the few Spanish-English geological dictionaries available. Even geologists who are fluent in Spanish often encounter local terminology oijerga that is unfamiliar. These terms, which have grown out of five centuries of mining tradition in Latin America, and frequently draw on native languages, usually cannot be found in standard dictionaries. There are, of course, many geological terms which can be recognized even by geologists who speak little or no Spanish. -

BIOVIA Materials Studio CANTERA
MATERIALS STUDIO CANTERA OVERVIEW KEY USES OF CANTERA A detailed knowledge of reaction kinetics is required in order to accurately model chemically reacting or combusting systems. Reactor Design, Optimization & Improvement These systems are complex, with multiple interdependent Cantera allows users to explore the effects of reactor geometry reaction pathways and competing processes involved. Reaction and operating conditions on processes to identify trade- kinetics simulations provide predictions for the concentration offs, determine reaction controllability and optimize existing of reactants and products under a specific set of reactor reactors. conditions and geometries. Identification & Quantification of Emissions BIOVIA Materials Studio Cantera is designed to support these calculations by linking thermodynamic input and reaction Reaction kinetics simulations may be deployed to understand step data with established Cantera reaction kinetics solvers. In the byproducts of reactions and predict how process changes addition, users can supplement existing reaction mechanism can affect the process further downstream. data with quantum mechanical transition state calculations, all on a firm, thermodynamically-consistent basis. Sensitivity Analysis Users of Cantera can better identify dominant chemistry BIOVIA Materials Studio Cantera enables scientists and engineers mechanisms and understand experimentally observed results to make predictions covering a diverse range of problems, to study and optimize the most promising pathways with including exhaust-gas emissions, ignition phenomena, engine further experiments or simulations. knock, flame-speed, flammability limits, catalytic lightoff and chemical conversion. Reaction kinetics simulations for issues in fuel cell development, batteries research, fuel reforming, THE BIOVIA MATERIALS STUDIO ADVANTAGE material deposition and plasma processing research can all BIOVIA Materials Studio Cantera is part of BIOVIA Materials be addressed using BIOVIA Materials Studio. -

Doctorat De L'université De Toulouse
THESETHESE En vue de l'obtention du DOCTORAT DE L’UNIVERSITÉ DE TOULOUSE Délivré par l'Université Toulouse III - Paul Sabatier Discipline ou spécialité : Chimie Quantique et Computationnelle Présentée et soutenue par Marco Verdicchio Le 17/12/2012 Titre : Molecular simulations as test beds for bridging High Throughput and High Performance computing JURY Prof. Laganà Antonio Prof. Evangelisti Stefano Prof. Coletti Cecilia Prof. Alikhani Emsail Ecole doctorale : Ecole Doctorale Sciences de la Matiere Unité de recherche : Laboratoire de Chimie et Physique Quantiques - UMR5626 Directeur(s) de Thèse : Laganà Antonio, Evangelisti Stefano, Thierry Leininger Rapporteurs : Contents French summary 1 Introduction 21 1 Molecular Simulators 25 1.1 The general Schrodinger¨ equation . 25 1.2 The electronic structure . 26 1.3 The nuclei dynamics . 30 1.4 The chemical kinetics systems . 32 1.5 GEMS the GRID simulator . 35 1.6 The kinetics version of the workflow . 38 1.7 Bridging HTC to HPC . 39 2 Code interoperability in Quantum chemistry and Dynamics 45 2.1 Data of Ab initio Quantum Chemistry . 46 2.2 Data of Quantum Dynamics . 48 2.3 The data model: Q5cost . 49 2.4 The D5cost data model . 52 2.5 The Library . 53 2.6 Tools and Wrappers . 54 2.7 Performance and benchmarks . 57 3 High level ab initio calculations of small systems 61 3.1 Coupled-Cluster study: the electronic wavefunction of tetra- hedral clusters . 61 3.2 Geometry optimizations of the tetrahedral clusters . 68 iii CONTENTS 3.3 No-pair bonding: Cu4 basis sets and correlated orbitals . 75 3.4 The no-pair bond state . -

Sustained Petascale in Action: Enabling Transformative Research 2019 Annual Report Sustained Petascale in Action: Enabling Transformative Research 2019 Annual Report
SUSTAINED PETASCALE IN ACTION: ENABLING TRANSFORMATIVE RESEARCH 2019 ANNUAL REPORT SUSTAINED PETASCALE IN ACTION: ENABLING TRANSFORMATIVE RESEARCH 2019 ANNUAL REPORT Editor Catherine Watkins Creative Director Steve Duensing Graphic Designer Megan Janeski Project Director William Kramer The research highlighted in this book is part of the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) and the state of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana–Champaign and its National Center for Supercomputing Applications. Visit https://bluewaters.ncsa.illinois.edu/science-teams for the latest on Blue Waters- enabled science and to watch the 2019 Blue Waters Symposium presentations. CLASSIFICATION KEY To provide an overview of how science teams are using Blue Waters, researchers were asked if their work fit any of the following classifications (number responding in parentheses): Data-intensive: Uses large numbers of files, large disk space/ DI bandwidth, or automated workflows/offsite transfers, etc. (83) GPU-accelerated: Written to run faster on XK nodes than on GA XE nodes (55) LEADING BY EXAMPLE Thousand-node: Scales to at least 1,000 nodes for production TN science (74) Memory-intensive: Uses at least 50 percent of available memory MI on 1,000-node run (21) BW Only on Blue Waters: Research only possible on Blue Waters (43) The National Center for Super- Earlier in 2020 I was privileged to be part of a del- computing Applications (NCSA) egation of University of Illinois leaders that partici- Multiphysics/multiscale: Job spans multiple length/time scales or was funded in 1986 to enable dis- pated in discussions on the next frontier of AI, data MP physical/chemical processes (59) coveries not possible anywhere science, and design thinking with Illinois alumnus TABLE OF else. -

Construction, Analysis, and Modeling of Complex Reaction Systems Using
Construction, analysis, and modeling of complex reaction networks with RING A DISSERTATION SUBMITTED TO THE FACULTY OF THE GRADUATE SCHOOL OF THE UNIVERSITY OF MINNESOTA BY Srinivas Rangarajan IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Doctor of Philosophy Advised by Prodromos Daoutidis and Aditya Bhan August 2013 c Srinivas Rangarajan 2013 ALL RIGHTS RESERVED Acknowledgments I would like to thank my advisors Profs. Prodromos Daoutidis and Aditya Bhan. They gave me enough freedom to pursue my ideas, encouraged collaborations, offered inter- esting problems to work on, and above all were always available for advice to sort out my problems, technical and otherwise. I thank the members of the Daoutidis group, with whom I shared my office, and the Bhan group, with whom I had valuable interaction time-to-time. I would like to specifically thank my colleagues in the Daoutidis group, Ana Torres, Alex Marvin, Dimitrios Georgis, Seongmin Heo, Adam Kelloway, and former members Dr. Sujit Jogwar, Prof. Fernando Lima, and Prof. Milana Trifkovic, with whom I have had innumerable engaging technical discussions on common research problems. Beyond work, I have also found good friends in them. The camaraderie in the group has been wonderful and has made our office, the Popcorn Lounge, the ideal place to work; I will forever cherish the 2 PM coffee breaks. I also want to thank my batchmates and friends in the department, especially, Dr. Reetam Chakrabarty, with whom I had many valuable discussions starting from my first few weeks in Minneapolis. I thank all my other friends that I have made and all the roommates I have had here in the last five years { life in Minneapolis would never have been the cherishable if not for them. -

Easybuild Documentation Release 20170109.01
EasyBuild Documentation Release 20170109.01 Ghent University Fri, 27 Jan 2017 16:57:55 Contents 1 Introductory topics 3 1.1 What is EasyBuild?.........................................3 1.2 Concepts and terminology......................................4 1.2.1 EasyBuild framework....................................4 1.2.2 Easyblocks.........................................4 1.2.3 Toolchains..........................................5 1.2.4 Easyconfig files.......................................6 1.2.5 Extensions..........................................6 1.3 Typical workflow example: building and installing WRF......................6 1.3.1 Searching for available easyconfigs files..........................7 1.3.2 Getting an overview of planned installations........................7 1.3.3 Installing a software stack.................................8 2 Getting started 9 2.1 Installing EasyBuild.........................................9 2.1.1 Requirements........................................9 2.1.2 Bootstrapping EasyBuild.................................. 10 2.1.3 Advanced bootstrapping options.............................. 13 2.1.4 Updating an existing EasyBuild installation........................ 14 2.1.5 Dependencies........................................ 15 2.1.6 Sources........................................... 17 2.1.7 In case of installation issues.................................. 17 2.2 Configuring EasyBuild........................................ 18 2.2.1 Supported configuration types............................... 18 2.2.2 Overview