2011: Large-Scale Combustion Simulations with Detailed Reaction Chemistry PI: Ingmar M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(12) United States Patent (10) Patent No.: US 6,492,429 B1 Graus Et Al
USOO6492429B1 (12) United States Patent (10) Patent No.: US 6,492,429 B1 Graus et al. (45) Date of Patent: Dec. 10, 2002 (54) COMPOSITION FOR THE TREATMENT OF 5,401.777 A 3/1995 Ammon et al. OSTEOARTHRITIS 5,494.668 A 2/1996 Patwardhan 5,629,351 A 5/1997 Taneja et al. (75) Inventors: Ivo Maria Franciscus Graus, Wg Ede 5,872,124. A * 2/1999 Koprowski et al. ......... 514/261 (NL); Hobbe Friso Smit, As Utrecht 5,888,514 A * 3/1999 Weisman ................. 424/195.1 (NL) FOREIGN PATENT DOCUMENTS (73) Assignee: N.V. Nutricia, Zoetermeer (NL) WO 95 22323 8/1995 WO 97 O7796 3/1997 (*) Notice: Subject to any disclaimer, the term of this OTHER PUBLICATIONS patent is extended or adjusted under 35 U.S.C. 154(b) by 0 days. Balch et al. Prescription for Nutritional Healing; Avery Publishing 2nd Ed. pp. 138–144, Oct. 1997.* (21) Appl. No.: 09/662,123 Lafeber et al., “Apocynin, a plant-derived, cartilage-Saving drug, might be useful in the treatment of rheumatoid (22) Filed: Sep. 14, 2000 arthritis", Rheumatology, (1999), pp. 1088–1093, vol. 38, O O British Society for Rheumatology. Related U.S. Application Data * cited by examiner (63) Continuation of application No. 09/613,562, filed on Jul. 10, 2000. Primary Examiner-Christopher R. Tate 7 ASSistant Examiner Patricia A Patten (51) Int. Cl." ......................... A01N 35/00; AO1N 65/00 (74) Attorney, Agent, or Firm-Browdy and Neimark, (52) U.S. Cl. ........................................ 514/688; 424/725 PL.L.C. (58) Field of Search .......................... -

Table S1. GC-MS Analysis of the Chloroform Soluble Fraction of Chestnut Wood Extractives
Table S1. GC-MS analysis of the chloroform soluble fraction of chestnut wood extractives. Untreated THM (chloroform soluble (chloroform soluble fraction) fraction) Extraction technique Extraction techniques a a b W/POM ASE E/T Water ASE E/T Water E/T/M c r.t. Area [%] Area [%] Compound KI [min] Furan 4.04 492 0.23 2-Furancarboxylic Acid 4.28 836 0.36 0.36 0.12 2.18 Benzaldehyde 4.31 961 0.29 1.34 1.49 Phenol 4.47 967 0.05 0.05 Methyl 2-Furancarboxylate 5.17 985 0.10 Benzyl Alcohol 5.21 1007 0.34 0.59 Levulinic Acid 5.34 1063 0.10 0.10 9.91 p-Cresol 5.61 1077 0.02 0.07 PhenylmethylFormate 5.66 1082 0.13 Nonanal 5.80 1105 0.08 0.04 2-Methoxyphenol 5.82 1106 5.88 Methyl Benzoate 5.88 1111 0.09 0.10 Benzaldehyde Dimethyl Acetal 6.03 1200 0.11 Maltol 6.05 1140 0.11 0.11 PhenylmethylAcetate 6.54 1162 0.10 Creosol 6.84 1203 0.06 5-Hydroxymethylfurfural 7.15 1224 3.60 0.90 NonanoicAcid 7.40 1272 0.05 3.60 2,3-Dihydro-3,5-Dihydroxy-6- Methyl-4H- 7.78 1290 Pyran-4-One 5-Acetoxymethyl-2-Furaldehyde 7.94 1304 0.27 2,6-Decadienal 8.04 1317 0.04 2-Methoxy-4-Vinylphenol 8.06 1320 0.11 0.20 2,6-Dimetyhoxyphenol 8.14 1357 0.14 0.17 0.17 0.17 4.16 DecanoicAcid 8.16 1370 0.07 0.13 2-Methoxy-4-Propylphenol 8.18 1382 0.19 0.34 0.80 Eugenol 8.20 1389 0.13 0.13 0.15 (e)-2-Tetradecene 8.37 1421 0.66 2.43 0.37 2,2'-Dimethylbiphenyl 8.44 1425 (e)-Cinnamic Acid 8.67 1430 0.05 Methyl 2- 8.78 1433 0.18 Phenylcyclopropancarboxylate 1-Methyl-3-(1-Methyl-2- 8.80 1435 0.38 Propenyl)Benzene 2,3-Dihydro-5,6-Dimethyl-1H-Indene 8.82 1438 0.09 Vanillin 8.95 1440 0.34 0.16 -

BOOK of ABSTRACTS Twelfth International Undergraduate Summer Research Symposium Thursday, August 1, 2019
BOOK OF ABSTRACTS Twelfth International Undergraduate Summer Research Symposium Thursday, August 1, 2019 Copyright © 2019 by New Jersey Institute of Technology (NJIT). All rights reserved. New Jersey Institute of Technology University Heights Newark, NJ 07102-1982 Joel S. Bloom President August 1, 2019 Welcome all – students, faculty, industry mentors, sponsors and friends of the university – to NJIT’s Twelfth International Undergraduate Summer Research Symposium. It is exciting to see so many ingenious inventions, and the bright, enterprising minds behind them, gathered in one place. That some of you have joined our innovation hub from as far away as India is a testament to the power of collaboration in the service of progress – not just in our own state or country, but across the globe. I want to especially thank the Provost’s office for making undergraduate research a high priority on our campus, the students’ advisers for their ideas and precious time over the summer, and our many sponsors for their generosity and commitment to helping forge the problem-solvers of tomorrow - today. And to the more than 130 of you exhibiting your work at the symposium, congratulations! By thinking creatively, following through with diligence and tenacity – and even retooling when the evidence requires it – you have embraced the rigors of professional science. You make us proud, and we look forward to following your successes in the years to come. Sincerely, Joel S. Bloom President Fadi P. Deek Provost and Senior Executive Vice President August 1, 2019 A message from the Provost: Welcome to NJIT’s Twelfth International Undergraduate Summer Research Symposium. -

Efeito Dos Metóxi-Catecóis Apocinina, Curcumina E Vanilina
i RESUMO O tamoxifeno (TAM) é um agente sintético, anti-estrogênico e não esteroidal, que comumente é prescrito no tratamento de pacientes com câncer de mama. Vários efeitos colaterais estão associados ao seu uso, como alterações vaginais, irregularidade menstrual, formação de pólipos no endométrio, cistos ovarianos, tromboembolismo, hepatocarcinoma, entre outros. Alguns produtos naturais são uma excelente estratégia na busca de novas drogas antitumorais, devido ao conhecimento popular de seu uso. A associacão de produtos naturais às drogas quimioterápicas clássicas tem mostrado um efeito sinérgico de grande interesse para a terapia antitumoral. A atividade citotóxica da curcumina é bem estabelecida em vários tipos de linhagens de células tumorais e tem sido amplamente estudada. A vanilina também tem mostrado atividade sobre as células tumorais devido aos seus efeitos citotóxicos e citostáticos. Já os efeitos da apocinina são devidos, principalmente, à sua eficiente inibição do complexo NADPH-oxidase, e consequentemente, de espécies reativas de oxigênio. O objetivo deste trabalho foi avaliar o efeito dos metóxi-catecóis apocinina, curcumina e vanilina sobre a citotoxicidade em células normais, hemácias e leucócitos polimorfonucleares, e em células de leucemia mielóide crônica humana (K562) exercida pelo TAM, como também a atividade antioxidante destes compostos. A citotoxidade foi analisada em hemácias através da liberação de hemoglobina e K+, e a curcumina foi o único composto que diminuiu a citotoxicidade do TAM sobre essas células. -

Package Name Software Description Project
A S T 1 Package Name Software Description Project URL 2 Autoconf An extensible package of M4 macros that produce shell scripts to automatically configure software source code packages https://www.gnu.org/software/autoconf/ 3 Automake www.gnu.org/software/automake 4 Libtool www.gnu.org/software/libtool 5 bamtools BamTools: a C++ API for reading/writing BAM files. https://github.com/pezmaster31/bamtools 6 Biopython (Python module) Biopython is a set of freely available tools for biological computation written in Python by an international team of developers www.biopython.org/ 7 blas The BLAS (Basic Linear Algebra Subprograms) are routines that provide standard building blocks for performing basic vector and matrix operations. http://www.netlib.org/blas/ 8 boost Boost provides free peer-reviewed portable C++ source libraries. http://www.boost.org 9 CMake Cross-platform, open-source build system. CMake is a family of tools designed to build, test and package software http://www.cmake.org/ 10 Cython (Python module) The Cython compiler for writing C extensions for the Python language https://www.python.org/ 11 Doxygen http://www.doxygen.org/ FFmpeg is the leading multimedia framework, able to decode, encode, transcode, mux, demux, stream, filter and play pretty much anything that humans and machines have created. It supports the most obscure ancient formats up to the cutting edge. No matter if they were designed by some standards 12 ffmpeg committee, the community or a corporation. https://www.ffmpeg.org FFTW is a C subroutine library for computing the discrete Fourier transform (DFT) in one or more dimensions, of arbitrary input size, and of both real and 13 fftw complex data (as well as of even/odd data, i.e. -

RMG-Py and Cantherm Documentation ⇌Release 2.0.0
RMG RMG-Py and CanTherm Documentation ⇌Release 2.0.0 William H. Green, Richard H. West, and the RMG Team Sep 16, 2016 CONTENTS 1 RMG User’s Guide 3 1.1 Introduction...............................................3 1.2 Release Notes..............................................4 1.3 Overview of Features...........................................6 1.4 Installation................................................7 1.5 Creating Input Files........................................... 20 1.6 Example Input Files........................................... 29 1.7 Running a Job.............................................. 36 1.8 Analyzing the Output Files........................................ 37 1.9 Species Representation.......................................... 38 1.10 Group Representation.......................................... 39 1.11 Databases................................................. 39 1.12 Thermochemistry Estimation...................................... 58 1.13 Kinetics Estimation........................................... 63 1.14 Liquid Phase Systems.......................................... 65 1.15 Guidelines for Building a Model..................................... 72 1.16 Standalone Modules........................................... 73 1.17 Frequently Asked Questions....................................... 81 1.18 Credits.................................................. 82 1.19 How to Cite................................................ 82 2 CanTherm User’s Guide 83 2.1 Introduction.............................................. -

Apocynin Exerts Dose-Dependent Cardioprotective Effects by Attenuating Reactive Oxygen Species in Ischemia/Reperfusion
maco har log P y: r O la Chen et al., Cardiovasc Pharm Open Access 2016, 5:2 u p c e n s a A DOI: 10.4176/2329-6607.1000176 v c o c i e d r s a s Open Access C Cardiovascular Pharmacology: ISSN: 2329-6607 Research Article OpenOpen Access Access Apocynin Exerts Dose-Dependent Cardioprotective Effects by Attenuating Reactive Oxygen Species in Ischemia/Reperfusion Qian Chen, Woodworth Parker C, Issachar Devine, Regina Ondrasik, Tsion Habtamu, Kyle D Bartol, Brendan Casey, Harsh Patel, William Chau, Tarah Kuhn, Robert Barsotti and Lindon Young* Department of Bio-Medical Sciences, Philadelphia College of Osteopathic Medicine, Philadelphia, USA Abstract Ischemia/reperfusion results in cardiac contractile dysfunction and cell death partly due to increased reactive oxygen species and decreased endothelial-derived nitric oxide bioavailability. NADPH oxidase normally produces reactive oxygen species to facilitate cell signalling and differentiation; however, excessive release of such species following ischemia exacerbates cell death. Thus, administration of an NADPH oxidase inhibitor, apocynin, may preserve cardiac function and reduce infarct size following ischemia. Apocynin dose-dependently (40 μM, 400 μM and 1 mM) attenuated leukocyte superoxide release by 87 ± 7%. Apocynin was also given to isolated perfused hearts after ischemia, with infarct size decreasing to 39 ± 7% (40 μM), 28 ± 4% (400 μM; p < 0.01) and 29 ± 6% (1 mM; p < 0.01), versus the control’s 46 ± 2%. This decrease correlated with improved final post-reperfusion left ventricular end-diastolic pressure, which decreased from 60 ± 5% in control hearts to 56 ± 5% (40 µM), 43 ± 4% (400 μM; p < 0.01) and 48 ± 5% (1 mM; p < 0.05), compared to baseline. -

Pyjac: Analytical Jacobian Generator for Chemical Kinetics
pyJac: analytical Jacobian generator for chemical kinetics Kyle E. Niemeyera,∗, Nicholas J. Curtisb, Chih-Jen Sungb aSchool of Mechanical, Industrial, and Manufacturing Engineering Oregon State University, Corvallis, OR 97331, USA bDepartment of Mechanical Engineering University of Connecticut, Storrs, CT, 06269, USA Abstract Accurate simulations of combustion phenomena require the use of detailed chemical kinet- ics in order to capture limit phenomena such as ignition and extinction as well as predict pollutant formation. However, the chemical kinetic models for hydrocarbon fuels of prac- tical interest typically have large numbers of species and reactions and exhibit high levels of mathematical stiffness in the governing differential equations, particularly for larger fuel molecules. In order to integrate the stiff equations governing chemical kinetics, generally reactive-flow simulations rely on implicit algorithms that require frequent Jacobian matrix evaluations. Some in situ and a posteriori computational diagnostics methods also require accurate Jacobian matrices, including computational singular perturbation and chemical ex- plosive mode analysis. Typically, finite differences numerically approximate these, but for larger chemical kinetic models this poses significant computational demands since the num- ber of chemical source term evaluations scales with the square of species count. Furthermore, existing analytical Jacobian tools do not optimize evaluations or support emerging SIMD processors such as GPUs. Here we introduce pyJac, a Python-based open-source program that generates analytical Jacobian matrices for use in chemical kinetics modeling and anal- ysis. In addition to producing the necessary customized source code for evaluating reaction rates (including all modern reaction rate formulations), the chemical source terms, and the Jacobian matrix, pyJac uses an optimized evaluation order to minimize computational and memory operations. -

Chapter 6: Effects of Apocynin and Analogs on the Cytokine
Chapter 6 Effects of apocynin and analogs on the cytokine production by mononuclear cells E. van den Worm#, M.E. Vianen*, C.J. Beukelman#, A.J.J. van den Berg#, R.P. Labadie#, H. van Dijk#,†, F.P.J.G. Lafeber*and J.W.J. Bijlsma*. # Department of Medicinal Chemistry, Utrecht Institute for Pharmaceutical Sciences, Faculty of Pharmacy, Utrecht University, PO Box 80082, 3508 TB Utrecht, The Netherlands. * Department of Rheumatology and Clinical Immunology, University Medical Center Utrecht, PO BOX 85500, 3508 GA Utrecht, The Netherlands. † Eijkman-Winkler Institute for Microbiology, Infectious Diseases and Inflammation, University Medical Center Utrecht, AZU G04.614, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands. - Je moet een vraag wel stellen zoals die gesteld hoort te worden - (Johan Cruijff, 1993) Chapter 6 Abstract Apocynin is a promising, plant-derived, nonsteroidal, anti-inflammatory compound that has been studied in different in vitro systems as well as in in vivo models for chronic inflammatory diseases. So far, apocynin is known as a specific inhibitor of NADPH-oxidase activity in stimulated neutrophils. This paper is dealing with possible other aspects of apocynin action, including inhibition of cytokine (IL-1, TNFα, IFNγ) production by cultured monocytes/macrophages and T cells, as well as inhibition of the proliferative response of T cells. Our findings implicate that not only apocynin itself, but also combinations of apocynin with one or more of its major in vivo metabolites may be involved in its net in vivo effect. This putative synergistic activity will be subject of further in vitro studies. 90 Effects of apocynin on cytokine production Introduction Apocynin has proven its potential value in the treatment of several experimental inflammatory diseases such as colitis (1), atherosclerosis (2), and rheumatoid arthritis (3). -
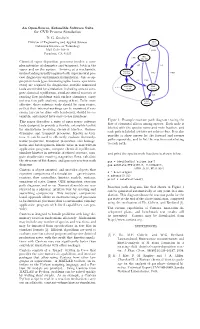
An Open-Source, Extensible Software Suite for CVD Process Simulation D
An Open-Source, Extensible Software Suite for CVD Process Simulation D. G. Goodwin Division of Engineering and Applied Science SIH4 0.0016 California Institute of Technology Mail Code 104-44 0.0135 Pasadena, CA 91125 SIH2 0.852 4.8e-005 Chemical vapor deposition processes involve a com- plex interplay of chemistry and transport, both in the 0.85 0.4 vapor and on the surface. Arriving at a mechanistic H3SISIH 0.415 0.429 understanding usually requires both experimental pro- 2.9e-005 cess diagnostics and numerical simulation. Just as ap- propriate tools (gas chromatographs, lasers, spectrom- 1 0.8 0.429 eters) are required for diagnostics, suitable numerical H2SISIH2 SI3H8 0.101 tools are needed for simulation, including ones to com- 0.0038 7.3e-005 pute chemical equilibrium, simulate stirred reactors or reacting flow problems with surface chemistry, carry 0.83 out reaction path analysis, among others. To be most SI2H6 effective, these software tools should be open-source, 0.00019 so that their internal workings can be examined if nec- essary (as can be done with hardware), should be ex- tensible, and should have easy-to-use interfaces. Figure 1: Example reaction path diagram tracing the This paper describes a suite of open-source software flow of elemental silicon among species. Each node is tools designed to provide a flexible, extensible toolkit labeled with the species name and mole fraction, and for simulations involving chemical kinetics, thermo- each path is labeled with its net relative flux. It is also dynamics, and transport processes. Known as Can- possible to show arrows for the forward and reverse tera, it can be used to efficiently evaluate thermody- paths separately, and to list the reaction contributing namic properties, transport properties, and homoge- to each path. -

BIOVIA MATERIALS STUDIO OVERVIEW Datasheet
BIOVIA MATERIALS STUDIO OVERVIEW Datasheet 10 Å 3 nm 15 µm Modeling and Simulation for Next Generation Materials. BIOVIA Materials Studio® is a complete modeling and simulation environment that enables researchers in materials science and chemistry to develop new materials by predicting the relationships of a material’s atomic and molecular structure with its properties and behavior. Using Materials Studio, researchers in many industries can engineer better performing materials of all types, including pharmaceuticals, catalysts, polymers and composites, metals and alloys, batteries and fuel cells, nanomaterials, and more. Materials Studio is the world’s most advanced, yet easy Materials Visualizer provides capability to construct, manipulate to use environment for modeling and evaluating materials and view models of molecules, crystalline materials, surfaces, performance and behavior. Using Materials Studio, materials polymers, and mesoscale structures. It also supports the full scientists experience the following benefits: range of Materials Studio simulations with capabilities to • Reduction in cost and time associated with physical testing visualize results through images, animations, graphs, charts, and experimentation through “Virtual Screening” of candidate tables, and textual data. Most tools in the Materials Visualizer material variations. can also be accessed through the MaterialsScript API, allowing • Acceleration of the innovation process - developing new, better expert users to create custom capabilities and automate performing, more sustainable, and cost effective materials faster repetitive tasks. Materials Visualizer’s Microsoft Windows than can be done with physical testing and experimentation. client operates with a range of Windows and Linux server • Improved fundamental understanding of the relationship architectures to provide a highly responsive user experience. between atomic and molecular structure with material properties and behavior. -

Quick Navigation Mechanical Engineering Department Facilities
Quick Navigation Department Facilities Multi-User Facilities Other Facilities Mechanical Engineering Department Facilities Departmental Facilities and Individual Faculty Research Laboratories: Shared Services Facilities Director: Haejune Kim We provide students and researchers open access to a wide range of equipment for measuring, characterizing and imaging of samples. If necessary, shared facility staff or super user of the instrument will offer training before giving access to the equipment. The shared services facilities are available to mechanical engineering faculty and students. Currently the group maintains and has available for scheduling the following machines:VEGA II LSU SEM, KLA- Tencor P-6 Stylus Profiler, VHX-600 Digital Microscope, High Performance Liquid Chromatography (HPLC), Total Organic Carbon (TOC) Analyzer, Atomic Layer Deposition (ALD) System, MicroClimate environmental chamber, Photron FSTCAM SA5 High Speed Camera, Olympus BX 61/Visitech QLC-100 Confocal Microscopy, Simultaneous Thermal Analyzer – Q600 SDT, Differential Scanning Calorimeter – DSC Q1000, Veeco Dektak 150 Profilometer, and Salt Spray Chamber. Acoustics and Signal Processing Laboratory Contact: Dr. Yong-Joe Kim The ASPL, founded in 2009, is a research laboratory of the Department of Mechanical Engineering at Texas A&M University (TAMU), College Station, Texas, USA. It is located at James J. Cain ’51 Building #409. Prof. Yong-Joe Kim currently serves as the director for the ASPL. The current research is focused on the areas of acoustics, signal processing, vibration, dynamics, and biomechanics. Advanced Computational Mechanics Laboratory Contact: Dr. J.N. Reddy Professor Reddy’s Advanced Computational Mechanics Laboratory (ACML) at Texas A&M University is dedicated to state-of-the-art research in the development of novel mathematical models and numerical simulation of physical phenomena.