WO 2017/075465 Al 4 May 20 17 (04.05.2017) W P O P C T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of the Transforming STRN-ALK Fusion As a Potential Therapeutic Target in the Aggressive Forms of Thyroid Cancer
Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer Lindsey M. Kellya,1, Guillermo Barilab,1, Pengyuan Liuc,1, Viktoria N. Evdokimovaa, Sumita Trivedid, Federica Panebiancoa, Manoj Gandhia, Sally E. Cartye, Steven P. Hodakf, Jianhua Luoa, Sanja Dacica, Yan P. Yua, Marina N. Nikiforovaa, Robert L. Ferrisd, Daniel L. Altschulerb, and Yuri E. Nikiforova,2 aDepartment of Pathology and Laboratory Medicine, bDepartment of Pharmacology and Chemical Biology, dDepartment of Otolaryngology, eDepartment of Surgery, Division of Endocrine Surgery, and fDepartment of Medicine, Division of Endocrinology and Metabolism, University of Pittsburgh School of Medicine, Pittsburgh, PA 15213; and cDepartment of Physiology and Cancer Center, Medical College of Wisconsin, Milwaukee, WI 53226 Edited* by Albert de la Chapelle, Ohio State University Comprehensive Cancer Center, Columbus, OH, and approved January 10, 2014 (received for review November 24, 2013) Thyroid cancer is a common endocrine malignancy that encom- However, a significant proportion of thyroid cancers have no passes well-differentiated as well as dedifferentiated cancer types. known driver mutations. The discovery of novel genetic events The latter tumors have high mortality and lack effective therapies. has been accelerated more recently due to the availability of next- Using a paired-end RNA-sequencing approach, we report the dis- generation sequencing approaches that allow investigators to ob- covery of rearrangements involving the anaplastic lymphoma tain information on the entire genome, exome, or transcriptome kinase (ALK) gene in thyroid cancer. The most common of these of tumor cells (6). In this study, we used whole-transcriptome involves a fusion between ALK and the striatin (STRN) gene, which [RNA-sequencing (RNA-Seq)] analysis to identify novel gene fu- is the result of a complex rearrangement involving the short arm sions in thyroid cancer. -

Molecular Genetic Analysis of Two Loci (Ity2 and Ity3) Involved in The
Copyright Ó 2007 by the Genetics Society of America DOI: 10.1534/genetics.107.075523 Molecular Genetic Analysis of Two Loci (Ity2 and Ity3) Involved in the Host Response to Infection With Salmonella Typhimurium Using Congenic Mice and Expression Profiling Vanessa Sancho-Shimizu,*,† Rabia Khan,*,† Serge Mostowy,† Line Larivie`re,† Rosalie Wilkinson,† Noe´mie Riendeau,† Marcel Behr† and Danielle Malo*,†,‡,1 *Department of Human Genetics, McGill University, Montreal, Quebec H3G 1A4, Canada and †Center for the Study of Host Resistance, McGill University Health Center, Montreal, Quebec H3G 1A4, Canada and ‡Department of Medicine, McGill University, Montreal, Quebec H3G 1A4, Canada Manuscript received May 4, 2007 Accepted for publication July 27, 2007 ABSTRACT Numerous genes have been identified to date that contribute to the host response to systemic Sal- monella Typhimurium infection in mice. We have previously identified two loci, Ity2 and Ity3, that control survival to Salmonella infection in the wild-derived inbred MOLF/Ei mouse using a (C57BL/6J 3 MOLF/ Ei)F2cross. We validated the existence of these two loci by creating congenic mice carrying each quan- titative trait locus (QTL) in isolation. Subcongenic mice generated for each locus allowed us to define the critical intervals underlying Ity2 and Ity3. Furthermore, expression profiling was carried out with the aim of identifying differentially expressed genes within the critical intervals as potential candidate genes. Genomewide expression arrays were used to interrogate expression differences in the Ity2 congenics, leading to the identification of a new candidate gene (Havcr2, hepatitis A virus cellular receptor 2). Interval-specific oligonucleotide arrays were created for Ity3, identifying one potential candidate gene (Chi3l1, chitinase 3-like 1) to be pursued further. -

A Genome‐Wide Association Study Highlights a Regulatory Role for IFNG‐AS1
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.13.903989; this version posted January 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A genome‐wide association study highlights a regulatory role for IFNG‐AS1 contributing to cutaneous leishmaniasis in Brazil Short title: A GWAS for cutaneous leishmaniasis in Brazil Léa C. Castellucci1,2,*, Lucas Almeida1,2,*, Svetlana Cherlin3,*, Michaela Fakiola4, Edgar Carvalho1, Amanda B. Figueiredo5, Clara M. Cavalcanti5, Natalia S. Alves5, Walderez O. Dutra1,6 , Kenneth J. Gollob1,5,7, Heather J. Cordell3 , and Jenefer M. Blackwell8,9 *Equal contributions 1National Institute of Science and Technology in Tropical Diseases, Brazil; 2Federal University of Bahia, Salvador, Brazil; 3Population Health Sciences Institute, Newcastle University, UK; 4INGM‐National Institute of Molecular Genetics "Romeo ed Enrica Invernizzi" Milan, Milan, Italy; 5International Center for Research, AC Camargo Cancer Center, São Paulo, Brazil; 6Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil; 7Núcleo de Ensino e Pesquisa, Instituto Mario Penna, Belo Horizonte, Brazil; 8Department of Pathology, University of Cambridge, UK; 9Telethon Kids Institute, The University of Western Australia, Western Australia Corresponding author: Jenefer M. Blackwell ([email protected] Correspondence to: Jenefer M. Blackwell, PO Box 855, West Perth, Western Australia 6872; Hospital Avenue, Nedlands, Western Australia 6009; Phone: +61 8 63191000. Funding British Medical Research Council (MRC) MR/N017390/1, FAPEMIG grant in cooperation with MRC/CONFAP (CBB-APQ-00883-16), National Institute of Science and Technology in Tropical Diseases, Brazil (N° 573839/2008–5), CNPq (K.J.G. -

Mediator of DNA Damage Checkpoint 1 (MDC1) Is a Novel Estrogen Receptor Co-Regulator in Invasive 6 Lobular Carcinoma of the Breast 7 8 Evelyn K
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.16.423142; this version posted December 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Running Title: MDC1 co-regulates ER in ILC 2 3 Research article 4 5 Mediator of DNA damage checkpoint 1 (MDC1) is a novel estrogen receptor co-regulator in invasive 6 lobular carcinoma of the breast 7 8 Evelyn K. Bordeaux1+, Joseph L. Sottnik1+, Sanjana Mehrotra1, Sarah E. Ferrara2, Andrew E. Goodspeed2,3, James 9 C. Costello2,3, Matthew J. Sikora1 10 11 +EKB and JLS contributed equally to this project. 12 13 Affiliations 14 1Dept. of Pathology, University of Colorado Anschutz Medical Campus 15 2Biostatistics and Bioinformatics Shared Resource, University of Colorado Comprehensive Cancer Center 16 3Dept. of Pharmacology, University of Colorado Anschutz Medical Campus 17 18 Corresponding author 19 Matthew J. Sikora, PhD.; Mail Stop 8104, Research Complex 1 South, Room 5117, 12801 E. 17th Ave.; Aurora, 20 CO 80045. Tel: (303)724-4301; Fax: (303)724-3712; email: [email protected]. Twitter: 21 @mjsikora 22 23 Authors' contributions 24 MJS conceived of the project. MJS, EKB, and JLS designed and performed experiments. JLS developed models 25 for the project. EKB, JLS, SM, and AEG contributed to data analysis and interpretation. SEF, AEG, and JCC 26 developed and performed informatics analyses. MJS wrote the draft manuscript; all authors read and revised the 27 manuscript and have read and approved of this version of the manuscript. -

The MAP4K4-STRIPAK Complex Promotes Growth and Tissue Invasion In
bioRxiv preprint doi: https://doi.org/10.1101/2021.05.07.442906; this version posted May 8, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 The MAP4K4-STRIPAK complex promotes growth and tissue invasion in 2 medulloblastoma 3 Jessica Migliavacca1, Buket Züllig1, Charles Capdeville1, Michael Grotzer2 and Martin Baumgartner1,* 4 1 Division of Oncology, Children’s Research Center, University Children’s Hospital Zürich, Zürich, 5 Switzerland 6 2 Division of Oncology, University Children’s Hospital Zürich, Zürich, Switzerland 7 8 *e-mail: [email protected] 9 10 Abstract 11 Proliferation and motility are mutually exclusive biological processes associated with cancer that depend 12 on precise control of upstream signaling pathways with overlapping functionalities. We find that STRN3 13 and STRN4 scaffold subunits of the STRIPAK complex interact with MAP4K4 for pathway regulation in 14 medulloblastoma. Disruption of the MAP4K4-STRIPAK complex impairs growth factor-induced 15 migration and tissue invasion and stalls YAP/TAZ target gene expression and oncogenic growth. The 16 migration promoting functions of the MAP4K4-STRIPAK complex involve the activation of novel PKCs 17 and the phosphorylation of the membrane targeting S157 residue of VASP through MAP4K4. The anti- 18 proliferative effect of complex disruption is associated with reduced YAP/TAZ target gene expression 19 and results in repressed tumor growth in the brain tissue. This dichotomous functionality of the STRIPAK 20 complex in migration and proliferation control acts through MAP4K4 regulation in tumor cells and 21 provides relevant mechanistic insights into novel tumorigenic functions of the STRIPAK complex in 22 medulloblastoma. -

Detection of Aneuploidies by Paralogous Sequence Quantification S Deutsch, U Choudhury, G Merla, C Howald, a Sylvan, S E Antonarakis
908 J Med Genet: first published as 10.1136/jmg.2004.023184 on 9 December 2004. Downloaded from ORIGINAL ARTICLE Detection of aneuploidies by paralogous sequence quantification S Deutsch, U Choudhury, G Merla, C Howald, A Sylvan, S E Antonarakis ............................................................................................................................... J Med Genet 2004;41:908–915. doi: 10.1136/jmg.2004.023184 Background: Chromosomal aneuploidies are a common cause of congenital disorders associated with cognitive impairment and multiple dysmorphic features. Pre-natal diagnosis of aneuploidies is most See end of article for commonly performed by the karyotyping of fetal cells obtained by amniocentesis or chorionic villus authors’ affiliations sampling, but this method is labour intensive and requires about 14 days to complete. ....................... Methods: We have developed a PCR based method for the detection of targeted chromosome number Correspondence to: abnormalities termed paralogous sequence quantification (PSQ), based on the use of paralogous genes. Professor Stylianos E Paralogous sequences have a high degree of sequence identity, but accumulate nucleotide substitutions in Antonarakis, Department a locus specific manner. These sequence differences, which we term paralogous sequence mismatches of Genetic Medicine and Development, University of (PSMs), can be quantified using pyrosequencing technology, to estimate the relative dosage between Geneva Medical School, different chromosomes. We designed 10 assays for the detection of trisomies of chromosomes 13, 18, and GE 1211, Geneva, 21 and sex chromosome aneuploidies. Switzerland; Stylianos. antonarakis@medecine. Results: We evaluated the performance of this method on 175 DNAs, highly enriched for abnormal unige.ch samples. A correct and unambiguous diagnosis was given for 119 out of 120 aneuploid samples as well as for all the controls. -

Genomic and Expression Profiling of Chromosome 17 in Breast Cancer Reveals Complex Patterns of Alterations and Novel Candidate Genes
[CANCER RESEARCH 64, 6453–6460, September 15, 2004] Genomic and Expression Profiling of Chromosome 17 in Breast Cancer Reveals Complex Patterns of Alterations and Novel Candidate Genes Be´atrice Orsetti,1 Me´lanie Nugoli,1 Nathalie Cervera,1 Laurence Lasorsa,1 Paul Chuchana,1 Lisa Ursule,1 Catherine Nguyen,2 Richard Redon,3 Stanislas du Manoir,3 Carmen Rodriguez,1 and Charles Theillet1 1Ge´notypes et Phe´notypes Tumoraux, EMI229 INSERM/Universite´ Montpellier I, Montpellier, France; 2ERM 206 INSERM/Universite´ Aix-Marseille 2, Parc Scientifique de Luminy, Marseille cedex, France; and 3IGBMC, U596 INSERM/Universite´Louis Pasteur, Parc d’Innovation, Illkirch cedex, France ABSTRACT 17q12-q21 corresponding to the amplification of ERBB2 and collinear genes, and a large region at 17q23 (5, 6). A number of new candidate Chromosome 17 is severely rearranged in breast cancer. Whereas the oncogenes have been identified, among which GRB7 and TOP2A at short arm undergoes frequent losses, the long arm harbors complex 17q21 or RP6SKB1, TBX2, PPM1D, and MUL at 17q23 have drawn combinations of gains and losses. In this work we present a comprehensive study of quantitative anomalies at chromosome 17 by genomic array- most attention (6–10). Furthermore, DNA microarray studies have comparative genomic hybridization and of associated RNA expression revealed additional candidates, with some located outside current changes by cDNA arrays. We built a genomic array covering the entire regions of gains, thus suggesting the existence of additional amplicons chromosome at an average density of 1 clone per 0.5 Mb, and patterns of on 17q (8, 9). gains and losses were characterized in 30 breast cancer cell lines and 22 Our previous loss of heterozygosity mapping data pointed to the primary tumors. -

Plenary and Platform Abstracts
American Society of Human Genetics 68th Annual Meeting PLENARY AND PLATFORM ABSTRACTS Abstract #'s Tuesday, October 16, 5:30-6:50 pm: 4. Featured Plenary Abstract Session I Hall C #1-#4 Wednesday, October 17, 9:00-10:00 am, Concurrent Platform Session A: 6. Variant Insights from Large Population Datasets Ballroom 20A #5-#8 7. GWAS in Combined Cancer Phenotypes Ballroom 20BC #9-#12 8. Genome-wide Epigenomics and Non-coding Variants Ballroom 20D #13-#16 9. Clonal Mosaicism in Cancer, Alzheimer's Disease, and Healthy Room 6A #17-#20 Tissue 10. Genetics of Behavioral Traits and Diseases Room 6B #21-#24 11. New Frontiers in Computational Genomics Room 6C #25-#28 12. Bone and Muscle: Identifying Causal Genes Room 6D #29-#32 13. Precision Medicine Initiatives: Outcomes and Lessons Learned Room 6E #33-#36 14. Environmental Exposures in Human Traits Room 6F #37-#40 Wednesday, October 17, 4:15-5:45 pm, Concurrent Platform Session B: 24. Variant Interpretation Practices and Resources Ballroom 20A #41-#46 25. Integrated Variant Analysis in Cancer Genomics Ballroom 20BC #47-#52 26. Gene Discovery and Functional Models of Neurological Disorders Ballroom 20D #53-#58 27. Whole Exome and Whole Genome Associations Room 6A #59-#64 28. Sequencing-based Diagnostics for Newborns and Infants Room 6B #65-#70 29. Omics Studies in Alzheimer's Disease Room 6C #71-#76 30. Cardiac, Valvular, and Vascular Disorders Room 6D #77-#82 31. Natural Selection and Human Phenotypes Room 6E #83-#88 32. Genetics of Cardiometabolic Traits Room 6F #89-#94 Wednesday, October 17, 6:00-7:00 pm, Concurrent Platform Session C: 33. -

Novel Mutation and Three Other Sequence Variants Segregating with Phenotype at Keratoconus 13Q32 Susceptibility Locus
European Journal of Human Genetics (2012) 20, 389–397 & 2012 Macmillan Publishers Limited All rights reserved 1018-4813/12 www.nature.com/ejhg ARTICLE Novel mutation and three other sequence variants segregating with phenotype at keratoconus 13q32 susceptibility locus Marta Czugala1,6, Justyna A Karolak1,6, Dorota M Nowak1, Piotr Polakowski2, Jose Pitarque3, Andrea Molinari3, Malgorzata Rydzanicz1, Bassem A Bejjani4, Beatrice YJT Yue5, Jacek P Szaflik2 and Marzena Gajecka*,1 Keratoconus (KTCN), a non-inflammatory corneal disorder characterized by stromal thinning, represents a major cause of corneal transplantations. Genetic and environmental factors have a role in the etiology of this complex disease. Previously reported linkage analysis revealed that chromosomal region 13q32 is likely to contain causative gene(s) for familial KTCN. Consequently, we have chosen eight positional candidate genes in this region: MBNL1, IPO5, FARP1, RNF113B, STK24, DOCK9, ZIC5 and ZIC2, and sequenced all of them in 51 individuals from Ecuadorian KTCN families and 105 matching controls. The mutation screening identified one mutation and three sequence variants showing 100% segregation under a dominant model with KTCN phenotype in one large Ecuadorian family. These substitutions were found in three different genes: c.2262A4C (p.Gln754His) and c.720+43A4GinDOCK9; c.2377-132A4CinIPO5 and c.1053+29G4CinSTK24. PolyPhen analyses predicted that c.2262A4C (Gln754His) is possibly damaging for the protein function and structure. Our results suggest that c.2262A4C (p.Gln754His) -

Ck1δ Over-Expressing Mice Display ADHD-Like Behaviors, Frontostriatal Neuronal Abnormalities and Altered Expressions of ADHD-Candidate Genes
Molecular Psychiatry (2020) 25:3322–3336 https://doi.org/10.1038/s41380-018-0233-z ARTICLE CK1δ over-expressing mice display ADHD-like behaviors, frontostriatal neuronal abnormalities and altered expressions of ADHD-candidate genes 1 1 1 2 1 1 1 Mingming Zhou ● Jodi Gresack ● Jia Cheng ● Kunihiro Uryu ● Lars Brichta ● Paul Greengard ● Marc Flajolet Received: 8 November 2017 / Revised: 4 July 2018 / Accepted: 18 July 2018 / Published online: 19 October 2018 © Springer Nature Limited 2018 Abstract The cognitive mechanisms underlying attention-deficit hyperactivity disorder (ADHD), a highly heritable disorder with an array of candidate genes and unclear genetic architecture, remain poorly understood. We previously demonstrated that mice overexpressing CK1δ (CK1δ OE) in the forebrain show hyperactivity and ADHD-like pharmacological responses to D- amphetamine. Here, we demonstrate that CK1δ OE mice exhibit impaired visual attention and a lack of D-amphetamine- induced place preference, indicating a disruption of the dopamine-dependent reward pathway. We also demonstrate the presence of abnormalities in the frontostriatal circuitry, differences in synaptic ultra-structures by electron microscopy, as 1234567890();,: 1234567890();,: well as electrophysiological perturbations of both glutamatergic and GABAergic transmission, as observed by altered frequency and amplitude of mEPSCs and mIPSCs. Furthermore, gene expression profiling by next-generation sequencing alone, or in combination with bacTRAP technology to study specifically Drd1a versus Drd2 medium spiny neurons, revealed that developmental CK1δ OE alters transcriptional homeostasis in the striatum, including specific alterations in Drd1a versus Drd2 neurons. These results led us to perform a fine molecular characterization of targeted gene networks and pathway analysis. Importantly, a large fraction of 92 genes identified by GWAS studies as associated with ADHD in humans are significantly altered in our mouse model. -

Overexpressed WDR3 Induces the Activation of Hippo Pathway by Interacting with GATA4 in Pancreatic Cancer
Overexpressed WDR3 Induces the Activation of Hippo Pathway by Interacting with GATA4 in Pancreatic Cancer Wenjie Su Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Shikai Zhu Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Kai Chen Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Hongji Yang Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Mingwu Tian Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Qiang Fu Massachusetts General Hospital Ganggang Shi University of British Columbia School of Human Kinetics: The University of British Columbia School of Kinesiology Shijian Feng University of British Columbia School of Human Kinetics: The University of British Columbia School of Kinesiology Dianyun Ren Wuhan Union Hospital Xin Jin Wuhan Union Hospital Chong Yang ( [email protected] ) Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Page 1/34 Research Keywords: Pancreatic Cancer, WDR3, GATA4, YAP1, Hippo Signaling Pathway Posted Date: November 13th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-104564/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on March 1st, 2021. See the published version at https://doi.org/10.1186/s13046-021-01879-w. Page 2/34 Abstract Background: WD repeat domain 3 (WDR3) is involved in a variety of cellular processes including gene regulation, cell cycle progression, signal transduction and apoptosis. -
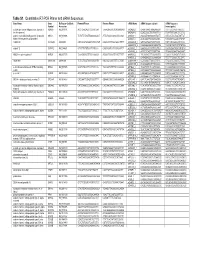
Table S1. Quantitative RT-PCR Primer and Sirna Sequences Gene Name
Table S1. Quantitative RT-PCR Primer and siRNA Sequences Gene Name Gene RefSeq or GenBank Forward Primer Reverse Primer siRNA Name siRNA Sequence (plus) siRNA Sequence Symbol Accession (minus/guide) a disintegrin and metalloproteinase domain 9 ADAM9 NM_003816 ACCTCAGCAGTTCCCATCAA TAAAGGAGGTGCAGGAGCAG siADAM9_1 GAGATTAACTAGAGAAAGA TCTTTCTCTAGTTAATCTC (meltrin gamma) siADAM9_2 GGAGGGAGTTCATAATTCA TGAATTATGAACTCCCTCC guanine nucleotide binding protein (G protein), GNAI3 NM_006496 TCTGTTATAGTTGGCGGCAGT ATTCTGCAAAGGCAAAAGGA siGNAI3_1 AGAACTAGCAGGAGTGATT AATCACTCCTGCTAGTTCT alpha inhibiting activity polypeptide 3 siGNAI3_2 ACACAGGTTCCAATACATA TATGTATTGGAACCTGTGT AA099748 AA099748 AA099748 CTACCACTGAGGTGTCCCTGT CAGCCATCTAACAGCATTTTT siAA099748_1 GTTAGATGGCTGATTAATA TATTAATCAGCCATCTAAC siAA099748_2 CGACAGAGGAGCAGCATTA TAATGCTGCTCCTCTGTCG calpain 12 CAPN12 NM_144691 GTCCTTCTGTCCCTCATCCA CAGCAGCTCCTCTGGAATCT siCAPN12_1 GGACACGCGTATTCCATCA TGATGGAATACGCGTGTCC siCAPN12_2 AATCCTCAGTTCCGTTTAA TTAAACGGAACTGAGGATT NMDA receptor regulated 1 NARG1 NM_057175 TAAAGGGAATTTGCCGAAGA AGCACTGGAGTTCGGTTTGT siNARG1_1 TGCGAGATCTTGAGGGTTA TAACCCTCAAGATCTCGCA siNARG1_2 GATAGGAGGTCCAAAAGAA TTCTTTTGGACCTCCTATC AK091308 AK091308 AK091308 TCTCCTGAATGGGAGGAATG GGCAACAGATGTTTCCTGTG siAK091308_1 CCAAAGACTTAGACTGTAA TTACAGTCTAAGTCTTTGG siAK091308_2 GCACAAAGCATTTACAACA TGTTGTAAATGCTTTGTGC serine/threonine kinase 24 (STE20 homolog, STK24 NM_003576 GCATCTGCCTTCCTTATCCA TGACAGTGTTTTGCCAGAGG siSTK24_1 TCGATTATCTCCATTCGGA TCCGAATGGAGATAATCGA yeast) siSTK24_2 CATCGGACTTGGACAGAAA