Novel Mutation and Three Other Sequence Variants Segregating with Phenotype at Keratoconus 13Q32 Susceptibility Locus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Rise and Fall of the Bovine Corpus Luteum
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 The Rise and Fall of the Bovine Corpus Luteum Heather Talbott University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, Molecular Biology Commons, and the Obstetrics and Gynecology Commons Recommended Citation Talbott, Heather, "The Rise and Fall of the Bovine Corpus Luteum" (2017). Theses & Dissertations. 207. https://digitalcommons.unmc.edu/etd/207 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. THE RISE AND FALL OF THE BOVINE CORPUS LUTEUM by Heather Talbott A DISSERTATION Presented to the Faculty of the University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor John S. Davis University of Nebraska Medical Center Omaha, Nebraska May, 2017 Supervisory Committee: Carol A. Casey, Ph.D. Andrea S. Cupp, Ph.D. Parmender P. Mehta, Ph.D. Justin L. Mott, Ph.D. i ACKNOWLEDGEMENTS This dissertation was supported by the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (NIFA) Pre-doctoral award; University of Nebraska Medical Center Graduate Student Assistantship; University of Nebraska Medical Center Exceptional Incoming Graduate Student Award; the VA Nebraska-Western Iowa Health Care System Department of Veterans Affairs; and The Olson Center for Women’s Health, Department of Obstetrics and Gynecology, Nebraska Medical Center. -

Exosomal Lncrna DOCK9-AS2 Derived from Cancer Stem Cell-Like
Dai et al. Cell Death and Disease (2020) 11:743 https://doi.org/10.1038/s41419-020-02827-w Cell Death & Disease ARTICLE Open Access Exosomal lncRNA DOCK9-AS2 derived from cancer stem cell-like cells activated Wnt/β-catenin pathway to aggravate stemness, proliferation, migration, and invasion in papillary thyroid carcinoma Wencheng Dai1, Xiaoxia Jin2,LiangHan1, Haijing Huang3,ZhenhuaJi1, Xinjiang Xu1, Mingming Tang1,BinJiang1 and Weixian Chen1 Abstract Exosomal long non-coding RNAs (lncRNAs) are crucial factors that mediate the extracellular communication in tumor microenvironment. DOCK9 antisense RNA2 (DOCK9-AS2) is an exosomal lncRNA which has not been investigated in papillary thyroid carcinoma (PTC). Based on the result of differentially expressed lncRNAs in PTC via bioinformatics databases, we discovered that DOCK9-AS2 was upregulated in PTC, and presented elevation in plasma exosomes of PTC patients. Functionally, DOCK9-AS2 knockdown reduced proliferation, migration, invasion, epithelial-to- mesenchymal (EMT) and stemness in PTC cells. PTC-CSCs transmitted exosomal DOCK9-AS2 to improve stemness of PTC cells. Mechanistically, DOCK9-AS2 interacted with SP1 to induce catenin beta 1 (CTNNB1) transcription and 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; sponged microRNA-1972 (miR-1972) to upregulate CTNNB1, thereby activating Wnt/β-catenin pathway in PTC cells. In conclusion, PTC-CSCs-derived exosomal lncRNA DOCK9-AS2 activated Wnt/β-catenin pathway to aggravate PTC progression, indicating that DOCK9-AS2 was a potential target for therapies in PTC. Introduction Therefore, more efforts are required for the improvement of Papillary thyroid cancer (PTC) takes up around 80% of targeted therapy and diagnosis in PTC. thyroid cancer (TC) cases1. Treatment outcome of PTC is Long non-coding RNAs (lncRNAs) are known as tran- generally satisfactory, and with appropriate treatment, over scripts without protein-coding ability and consist over 200 95% of PTC patients can survive longer than 5 years2. -

Transcriptome Analyses of Rhesus Monkey Pre-Implantation Embryos Reveal A
Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press Transcriptome analyses of rhesus monkey pre-implantation embryos reveal a reduced capacity for DNA double strand break (DSB) repair in primate oocytes and early embryos Xinyi Wang 1,3,4,5*, Denghui Liu 2,4*, Dajian He 1,3,4,5, Shengbao Suo 2,4, Xian Xia 2,4, Xiechao He1,3,6, Jing-Dong J. Han2#, Ping Zheng1,3,6# Running title: reduced DNA DSB repair in monkey early embryos Affiliations: 1 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China 2 Key Laboratory of Computational Biology, CAS Center for Excellence in Molecular Cell Science, Collaborative Innovation Center for Genetics and Developmental Biology, Chinese Academy of Sciences-Max Planck Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China 3 Yunnan Key Laboratory of Animal Reproduction, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China 4 University of Chinese Academy of Sciences, Beijing, China 5 Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China 6 Primate Research Center, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, 650223, China * Xinyi Wang and Denghui Liu contributed equally to this work 1 Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press # Correspondence: Jing-Dong J. Han, Email: [email protected]; Ping Zheng, Email: [email protected] Key words: rhesus monkey, pre-implantation embryo, DNA damage 2 Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press ABSTRACT Pre-implantation embryogenesis encompasses several critical events including genome reprogramming, zygotic genome activation (ZGA) and cell fate commitment. -

KETCH1 Imports HYL1 to Nucleus for Mirna Biogenesis in Arabidopsis
KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis Zhonghui Zhanga,b,1, Xinwei Guoa,c,1, Chunxiao Gea,1, Zeyang Maa, Mengqiu Jianga, Tianhong Lic, Hisashi Koiwad, Seong Wook Yange, and Xiuren Zhanga,2 aDepartment of Biochemistry and Biophysics, Institute for Plant Genomics and Biotechnology, Texas A&M University, College Station, TX 77843; bGuangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Science, South China Normal University, Guangzhou 510631, China; cCollege of Horticulture, China Agricultural University, Beijing 100193, China; dDepartment of Horticultural Sciences, Texas A&M University, College Station, TX 77843; and eDepartment of Systems Biology, College of Life Science and Biotechnology, Yonsei University, Seoul 120-749, Republic of Korea Edited by Xuemei Chen, University of California, Riverside, CA, and approved March 9, 2017 (received for review December 2, 2016) MicroRNA (miRNA) is processed from primary transcripts with hairpin premiRNAs in mammalians (11, 12). Importin-8 facilitates the structures (pri-miRNAs) by microprocessors in the nucleus. How recruitment of AGO2-containing RISC to target mRNAs to pro- cytoplasmic-borne microprocessor components are transported into mote efficient and specific gene silencing in the cytoplasm, whereas the nucleus to fulfill their functions remains poorly understood. Here, the protein can also transport AGO2 and AGO2 partners, GW we report KETCH1 (karyopherin enabling the transport of the proteins and miRNAs, into the nucleus to balance levels of cyto- cytoplasmic HYL1) as a partner of hyponastic leaves 1 (HYL1) protein, plasmic gene-silencing effectors (13–15). Arabidopsis encodes a core component of microprocessor in Arabidopsis and functional 18 importin β-proteins, among which few have also been reported counterpart of DGCR8/Pasha in animals. -

Detection of Aneuploidies by Paralogous Sequence Quantification S Deutsch, U Choudhury, G Merla, C Howald, a Sylvan, S E Antonarakis
908 J Med Genet: first published as 10.1136/jmg.2004.023184 on 9 December 2004. Downloaded from ORIGINAL ARTICLE Detection of aneuploidies by paralogous sequence quantification S Deutsch, U Choudhury, G Merla, C Howald, A Sylvan, S E Antonarakis ............................................................................................................................... J Med Genet 2004;41:908–915. doi: 10.1136/jmg.2004.023184 Background: Chromosomal aneuploidies are a common cause of congenital disorders associated with cognitive impairment and multiple dysmorphic features. Pre-natal diagnosis of aneuploidies is most See end of article for commonly performed by the karyotyping of fetal cells obtained by amniocentesis or chorionic villus authors’ affiliations sampling, but this method is labour intensive and requires about 14 days to complete. ....................... Methods: We have developed a PCR based method for the detection of targeted chromosome number Correspondence to: abnormalities termed paralogous sequence quantification (PSQ), based on the use of paralogous genes. Professor Stylianos E Paralogous sequences have a high degree of sequence identity, but accumulate nucleotide substitutions in Antonarakis, Department a locus specific manner. These sequence differences, which we term paralogous sequence mismatches of Genetic Medicine and Development, University of (PSMs), can be quantified using pyrosequencing technology, to estimate the relative dosage between Geneva Medical School, different chromosomes. We designed 10 assays for the detection of trisomies of chromosomes 13, 18, and GE 1211, Geneva, 21 and sex chromosome aneuploidies. Switzerland; Stylianos. antonarakis@medecine. Results: We evaluated the performance of this method on 175 DNAs, highly enriched for abnormal unige.ch samples. A correct and unambiguous diagnosis was given for 119 out of 120 aneuploid samples as well as for all the controls. -

Karyopherin Beta 3 (IPO5) (BD027479) Human Untagged Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC105419 Karyopherin beta 3 (IPO5) (BD027479) Human Untagged Clone Product data: Product Type: Expression Plasmids Product Name: Karyopherin beta 3 (IPO5) (BD027479) Human Untagged Clone Tag: Tag Free Symbol: IPO5 Vector: pCMV6-XL4 E. coli Selection: Ampicillin (100 ug/mL) Cell Selection: None Fully Sequenced ORF: >NCBI ORF sequence for BD027479, the custom clone sequence may differ by one or more nucleotides CTTCTCTCTCACGCCTAGCGCAATGGCGGCGGCCGCGGCGGASCAGCAACAGTTCTACCTGCTCCTGGGA AACCTGCTCAGCCCCGACAATGTGGTCCGGAAACAGGCAGAGGAAACCTATGAGAATANTCCCAGGCCAG TCAAAGATCACATTCCTCTTACAAGCCATCAGAAATACAACAGCTGCTGAAGAGGCTAGACAAATGGCCG CCGTTCTCCTAAGACGTCTCTTGTCCTCTGCATTTGATGAAGTCTATCCAGCACTTCCCTCTGATGTTCA GACTGCCATCAAGAGTGAGCTACTCATGATTATTCAGATGGAAACACAATCTAGCATGAGGAAAAAAGTT TGTGATATTGCGGSAGAACTGGCCAGGAATTTAATAGATGAGGATGGCAATAACCAGTGGCCCGAAGTTT GAAGTTCCTTTTTGATTCAGTCAG 5' Read Nucleotide >OriGene 5' read for BD027479 unedited Sequence: TGTATACGACTCATATAGGGCGGCCGCGAATCGGCACGAGGCGCCGGCGCCGGCGGCCGC GGCGGGGTGAGAGGCCGCGAGGCCCCGCCCCGTCCTCCCCTTTCCCCTTTGCCCCGCCCT TCCCGCGCGGCCCCCCGCAAGCCCCGCGCCGCCGCTGGTGCCGGTCCCCGCGCTGGGCCC GCCCCCGCCCCTCCCGCGGCCCGCGAGCGCGCCTCACGGCTCCTGTCTCCCCTCCCTCCT TCTCTCTCACGCCTAGCGCAATGGCGGCGGCCGCGGCGGAGCAGCAACAGTTCTACCTGC TCCTGGGAAACCTGCTCAGCCCCGACAATGTGGTCCGGAAACAGGCAGAGGAAACCTATG AGAATATCCCAGGCCAGTCAAAGATCACATTCCTCTTACAAGCCATCAGAAATACAACAG -

Genetic and Genomic Analysis of Hyperlipidemia, Obesity and Diabetes Using (C57BL/6J × TALLYHO/Jngj) F2 Mice
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Nutrition Publications and Other Works Nutrition 12-19-2010 Genetic and genomic analysis of hyperlipidemia, obesity and diabetes using (C57BL/6J × TALLYHO/JngJ) F2 mice Taryn P. Stewart Marshall University Hyoung Y. Kim University of Tennessee - Knoxville, [email protected] Arnold M. Saxton University of Tennessee - Knoxville, [email protected] Jung H. Kim Marshall University Follow this and additional works at: https://trace.tennessee.edu/utk_nutrpubs Part of the Animal Sciences Commons, and the Nutrition Commons Recommended Citation BMC Genomics 2010, 11:713 doi:10.1186/1471-2164-11-713 This Article is brought to you for free and open access by the Nutrition at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Nutrition Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Stewart et al. BMC Genomics 2010, 11:713 http://www.biomedcentral.com/1471-2164/11/713 RESEARCH ARTICLE Open Access Genetic and genomic analysis of hyperlipidemia, obesity and diabetes using (C57BL/6J × TALLYHO/JngJ) F2 mice Taryn P Stewart1, Hyoung Yon Kim2, Arnold M Saxton3, Jung Han Kim1* Abstract Background: Type 2 diabetes (T2D) is the most common form of diabetes in humans and is closely associated with dyslipidemia and obesity that magnifies the mortality and morbidity related to T2D. The genetic contribution to human T2D and related metabolic disorders is evident, and mostly follows polygenic inheritance. The TALLYHO/ JngJ (TH) mice are a polygenic model for T2D characterized by obesity, hyperinsulinemia, impaired glucose uptake and tolerance, hyperlipidemia, and hyperglycemia. -

A Rac/Cdc42 Exchange Factor Complex Promotes Formation of Lateral filopodia and Blood Vessel Lumen Morphogenesis
ARTICLE Received 1 Oct 2014 | Accepted 26 Apr 2015 | Published 1 Jul 2015 DOI: 10.1038/ncomms8286 OPEN A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis Sabu Abraham1,w,*, Margherita Scarcia2,w,*, Richard D. Bagshaw3,w,*, Kathryn McMahon2,w, Gary Grant2, Tracey Harvey2,w, Maggie Yeo1, Filomena O.G. Esteves2, Helene H. Thygesen2,w, Pamela F. Jones4, Valerie Speirs2, Andrew M. Hanby2, Peter J. Selby2, Mihaela Lorger2, T. Neil Dear4,w, Tony Pawson3,z, Christopher J. Marshall1 & Georgia Mavria2 During angiogenesis, Rho-GTPases influence endothelial cell migration and cell–cell adhesion; however it is not known whether they control formation of vessel lumens, which are essential for blood flow. Here, using an organotypic system that recapitulates distinct stages of VEGF-dependent angiogenesis, we show that lumen formation requires early cytoskeletal remodelling and lateral cell–cell contacts, mediated through the RAC1 guanine nucleotide exchange factor (GEF) DOCK4 (dedicator of cytokinesis 4). DOCK4 signalling is necessary for lateral filopodial protrusions and tubule remodelling prior to lumen formation, whereas proximal, tip filopodia persist in the absence of DOCK4. VEGF-dependent Rac activation via DOCK4 is necessary for CDC42 activation to signal filopodia formation and depends on the activation of RHOG through the RHOG GEF, SGEF. VEGF promotes interaction of DOCK4 with the CDC42 GEF DOCK9. These studies identify a novel Rho-family GTPase activation cascade for the formation of endothelial cell filopodial protrusions necessary for tubule remodelling, thereby influencing subsequent stages of lumen morphogenesis. 1 Institute of Cancer Research, Division of Cancer Biology, 237 Fulham Road, London SW3 6JB, UK. -

Genes with 5' Terminal Oligopyrimidine Tracts Preferentially Escape Global Suppression of Translation by the SARS-Cov-2 NSP1 Protein
Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Genes with 5′ terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 Nsp1 protein Shilpa Raoa, Ian Hoskinsa, Tori Tonna, P. Daniela Garciaa, Hakan Ozadama, Elif Sarinay Cenika, Can Cenika,1 a Department of Molecular Biosciences, University of Texas at Austin, Austin, TX 78712, USA 1Corresponding author: [email protected] Key words: SARS-CoV-2, Nsp1, MeTAFlow, translation, ribosome profiling, RNA-Seq, 5′ TOP, Ribo-Seq, gene expression 1 Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract Viruses rely on the host translation machinery to synthesize their own proteins. Consequently, they have evolved varied mechanisms to co-opt host translation for their survival. SARS-CoV-2 relies on a non-structural protein, Nsp1, for shutting down host translation. However, it is currently unknown how viral proteins and host factors critical for viral replication can escape a global shutdown of host translation. Here, using a novel FACS-based assay called MeTAFlow, we report a dose-dependent reduction in both nascent protein synthesis and mRNA abundance in cells expressing Nsp1. We perform RNA-Seq and matched ribosome profiling experiments to identify gene-specific changes both at the mRNA expression and translation level. We discover that a functionally-coherent subset of human genes are preferentially translated in the context of Nsp1 expression. These genes include the translation machinery components, RNA binding proteins, and others important for viral pathogenicity. Importantly, we uncovered a remarkable enrichment of 5′ terminal oligo-pyrimidine (TOP) tracts among preferentially translated genes. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -
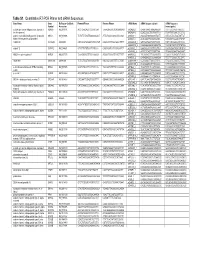
Table S1. Quantitative RT-PCR Primer and Sirna Sequences Gene Name
Table S1. Quantitative RT-PCR Primer and siRNA Sequences Gene Name Gene RefSeq or GenBank Forward Primer Reverse Primer siRNA Name siRNA Sequence (plus) siRNA Sequence Symbol Accession (minus/guide) a disintegrin and metalloproteinase domain 9 ADAM9 NM_003816 ACCTCAGCAGTTCCCATCAA TAAAGGAGGTGCAGGAGCAG siADAM9_1 GAGATTAACTAGAGAAAGA TCTTTCTCTAGTTAATCTC (meltrin gamma) siADAM9_2 GGAGGGAGTTCATAATTCA TGAATTATGAACTCCCTCC guanine nucleotide binding protein (G protein), GNAI3 NM_006496 TCTGTTATAGTTGGCGGCAGT ATTCTGCAAAGGCAAAAGGA siGNAI3_1 AGAACTAGCAGGAGTGATT AATCACTCCTGCTAGTTCT alpha inhibiting activity polypeptide 3 siGNAI3_2 ACACAGGTTCCAATACATA TATGTATTGGAACCTGTGT AA099748 AA099748 AA099748 CTACCACTGAGGTGTCCCTGT CAGCCATCTAACAGCATTTTT siAA099748_1 GTTAGATGGCTGATTAATA TATTAATCAGCCATCTAAC siAA099748_2 CGACAGAGGAGCAGCATTA TAATGCTGCTCCTCTGTCG calpain 12 CAPN12 NM_144691 GTCCTTCTGTCCCTCATCCA CAGCAGCTCCTCTGGAATCT siCAPN12_1 GGACACGCGTATTCCATCA TGATGGAATACGCGTGTCC siCAPN12_2 AATCCTCAGTTCCGTTTAA TTAAACGGAACTGAGGATT NMDA receptor regulated 1 NARG1 NM_057175 TAAAGGGAATTTGCCGAAGA AGCACTGGAGTTCGGTTTGT siNARG1_1 TGCGAGATCTTGAGGGTTA TAACCCTCAAGATCTCGCA siNARG1_2 GATAGGAGGTCCAAAAGAA TTCTTTTGGACCTCCTATC AK091308 AK091308 AK091308 TCTCCTGAATGGGAGGAATG GGCAACAGATGTTTCCTGTG siAK091308_1 CCAAAGACTTAGACTGTAA TTACAGTCTAAGTCTTTGG siAK091308_2 GCACAAAGCATTTACAACA TGTTGTAAATGCTTTGTGC serine/threonine kinase 24 (STE20 homolog, STK24 NM_003576 GCATCTGCCTTCCTTATCCA TGACAGTGTTTTGCCAGAGG siSTK24_1 TCGATTATCTCCATTCGGA TCCGAATGGAGATAATCGA yeast) siSTK24_2 CATCGGACTTGGACAGAAA -

Small-Molecule Inhibitors of Protein Acetylation and Deacetylation
Downloaded from rnajournal.cshlp.org on March 23, 2009 - Published by Cold Spring Harbor Laboratory Press Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation Andreas N. Kuhn, Maria A. van Santen, Andreas Schwienhorst, et al. RNA 2009 15: 153-175 originally published online November 24, 2008 Access the most recent version at doi:10.1261/rna.1332609 References This article cites 65 articles, 36 of which can be accessed free at: http://rnajournal.cshlp.org/content/15/1/153.full.html#ref-list-1 Email alerting Receive free email alerts when new articles cite this article - sign up in the box at the service top right corner of the article or click here To subscribe to RNA go to: http://rnajournal.cshlp.org/subscriptions Copyright © 2009 RNA Society JOBNAME: RNA 15#1 2009 PAGE: 1 OUTPUT: Tuesday December 9 15:25:03 2008 csh/RNA/175400/rna13326 Downloaded from rnajournal.cshlp.org on March 23, 2009 - Published by Cold Spring Harbor Laboratory Press Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation ANDREAS N. KUHN,1 MARIA A. VAN SANTEN,1 ANDREAS SCHWIENHORST,2 HENNING URLAUB,3 and REINHARD LU¨ HRMANN1 1Department of Cellular Biochemistry, Max Planck Institute for Biophysical Chemistry, D-37077 Go¨ttingen, Germany 2Department of Molecular Genetics and Preparative Molecular Biology, Institute for Microbiology and Genetics, D-37077 Go¨ttingen, Germany 3Bioanalytical Mass Spectrometry Group, Max Planck Institute for Biophysical Chemistry, D-37077 Go¨ttingen, Germany ABSTRACT The removal of intervening sequences from a primary RNA transcript is catalyzed by the spliceosome, a large complex consisting of five small nuclear (sn) RNAs and more than 150 proteins.