Prokaryotic Cell Structure and Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Review Pili in Gram-Negative and Gram-Positive Bacteria – Structure
Cell. Mol. Life Sci. 66 (2009) 613 – 635 1420-682X/09/040613-23 Cellular and Molecular Life Sciences DOI 10.1007/s00018-008-8477-4 Birkhuser Verlag, Basel, 2008 Review Pili in Gram-negative and Gram-positive bacteria – structure, assembly and their role in disease T. Profta,c,* and E. N. Bakerb,c a School of Medical Sciences, Department of Molecular Medicine & Pathology, University of Auckland, Private Bag 92019, Auckland 1142 (New Zealand), Fax: +64-9-373-7492, e-mail: [email protected] b School of Biological Sciences, University of Auckland, Auckland (New Zealand) c Maurice Wilkins Centre for Molecular Biodiscovery, University of Auckland (New Zealand) Received 08 August 2008; received after revision 24 September 2008; accepted 01 October 2008 Online First 27 October 2008 Abstract. Many bacterial species possess long fila- special form of bacterial cell movement, known as mentous structures known as pili or fimbriae extend- twitching motility. In contrast, the more recently ing from their surfaces. Despite the diversity in pilus discovered pili in Gram-positive bacteria are formed structure and biogenesis, pili in Gram-negative bac- by covalent polymerization of pilin subunits in a teria are typically formed by non-covalent homopo- process that requires a dedicated sortase enzyme. lymerization of major pilus subunit proteins (pilins), Minor pilins are added to the fiber and play a major which generates the pilus shaft. Additional pilins may role in host cell colonization. be added to the fiber and often function as host cell This review gives an overview of the structure, adhesins. Some pili are also involved in biofilm assembly and function of the best-characterized pili formation, phage transduction, DNA uptake and a of both Gram-negative and Gram-positive bacteria. -

Prokaryotes (Domains Bacteria & Archaea)
2/4/15 Prokaryotes (Domains Bacteria & Archaea) KEY POINTS 1. Decomposers: recycle organic and inorganic molecules in environment; makes them available to other organisms. 2. Essential components of symbioses. 3. Encompasses the origins of metabolism and metabolic diversity. 4. Origin of photosynthesis and formation of atmospheric Oxygen Ceno- Meso- zoic zoic ANTIQUITY Humans Paleozoic Colonization of land Animals Origin of solar system and Earth • >3.5 BILLION years old. • Alone for 2 1 4 billion years Proterozoic Archaean Prokaryotes Billions of 2 years ago3 Multicellular eukaryotes Single-celled eukaryotes Atmospheric oxygen General characteristics 1. Small: compare to 10-100µm for 0.5-5µm eukaryotic cell; single-celled; may form colonies. 2. Lack membrane- enclosed organelles. 3. Cell wall present, but different from plant cell wall. 1 2/4/15 General characteristics 4. Occur everywhere, most numerous organisms. – More individuals in a handful of soil then there are people that have ever lived. – By far more individuals in our gut than eukaryotic cells that are actually us. General characteristics 5. Metabolic diversity established nutritional modes of eukaryotes. General characteristics 6. Important decomposers and recyclers 2 2/4/15 General characteristics 6. Important decomposers and recyclers • Form the basis of global nutrient cycles. General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • About 1/2 of all human diseases are caused by Bacteria General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • Extremely important in agriculture as well. Pierce’s disease is caused by Xylella fastidiosa, a Gamma Proteobacteria. It causes over $56 million in damage annually in California. That’s with $34 million spent to control it! = $90 million in California alone. -

A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection
Published online: 2020-04-12 THIEME 302 A Champion of Host Defense: A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection Martin J. Page, BSc (Hons)1 Etheresia Pretorius, PhD1 1 Department of Physiological Sciences, Stellenbosch University, Address for correspondence Etheresia Pretorius, PhD, Department of Stellenbosch, South Africa Physiological Sciences, Faculty of Science, Stellenbosch University, Private Bag X1 Matieland, Stellenbosch, 7602, South Africa Semin Thromb Hemost 2020;46:302–319. (e-mail: [email protected]). Abstract Thrombocytopenia is commonly associated with sepsis and infections, which in turn are characterized by a profound immune reaction to the invading pathogen. Platelets are one of the cellular entities that exert considerable immune, antibacterial, and antiviral actions, and are therefore active participants in the host response. Platelets are sensitive to surrounding inflammatory stimuli and contribute to the immune response by multiple mechanisms, including endowing the endothelium with a proinflammatory phenotype, enhancing and amplifying leukocyte recruitment and inflammation, promoting the effector functions of immune cells, and ensuring an optimal adaptive immune response. During infection, pathogens and their products influence the platelet response and can even be toxic. However, platelets are able to sense and engage bacteria and viruses to assist in their removal and destruction. Platelets greatly contribute to host defense by multiple mechanisms, including forming immune complexes and aggregates, shedding their granular content, and internalizing pathogens and subsequently being marked for removal. These processes, and the nature of platelet function in general, cause the platelet to be irreversibly consumed in Keywords the execution of its duty. An exaggerated systemic inflammatory response to infection ► platelets can drive platelet dysfunction, where platelets are inappropriately activated and face ► virus immunological destruction. -

Killing of Gram-Negative Bacteria by Lactoferrin and Lysozyme
Killing of gram-negative bacteria by lactoferrin and lysozyme. R T Ellison 3rd, T J Giehl J Clin Invest. 1991;88(4):1080-1091. https://doi.org/10.1172/JCI115407. Research Article Although lactoferrin has antimicrobial activity, its mechanism of action is not full defined. Recently we have shown that the protein alters the Gram-negative outer membrane. As this membrane protects Gram-negative cells from lysozyme, we have studied whether lactoferrin's membrane effect could enhance the antibacterial activity of lysozyme. We have found that while each protein alone is bacteriostatic, together they can be bactericidal for strains of V. cholerae, S. typhimurium, and E. coli. The bactericidal effect is dose dependent, blocked by iron saturation of lactoferrin, and inhibited by high calcium levels, although lactoferrin does not chelate calcium. Using differing media, the effect of lactoferrin and lysozyme can be partially or completely inhibited; the degree of inhibition correlating with media osmolarity. Transmission electron microscopy shows that E. coli cells exposed to lactoferrin and lysozyme at 40 mOsm become enlarged and hypodense, suggesting killing through osmotic damage. Dialysis chamber studies indicate that bacterial killing requires direct contact with lactoferrin, and work with purified LPS suggests that this relates to direct LPS-binding by the protein. As lactoferrin and lysozyme are present together in high levels in mucosal secretions and neutrophil granules, it is probable that their interaction contributes to host defense. Find the latest version: https://jci.me/115407/pdf Killing of Gram-negative Bacteria by Lactofernn and Lysozyme Richard T. Ellison III*" and Theodore J. Giehl *Medical and tResearch Services, Department of Veterans Affairs Medical Center, and Division ofInfectious Diseases, Department ofMedicine, University ofColorado School ofMedicine, Denver, Colorado 80220 Abstract (4). -

Engineering the Human Microbiome, a Natural Double-Barreled Approach Towards Solving Both Our Current Antibiotic Resistance and Sepsis Problems
American Journal of www.biomedgrid.com Biomedical Science & Research ISSN: 2642-1747 --------------------------------------------------------------------------------------------------------------------------------- Opinion Copy Right@ Toleman MA Engineering the Human Microbiome, A Natural Double-Barreled Approach Towards Solving both our Current Antibiotic Resistance and Sepsis Problems Martins W1, Mathias J1, Babenko D2 and Toleman MA1* 1Cardiff University Medical school, Department of Immunology and Infection, The Heath hospital, UK 2Karaganda Medical University, Kazakhstan *Corresponding author: Toleman MA, Cardiff University Medical school, Department of Immunology and Infection, The Heath hospital, Cardiff, UK. To Cite This Article: Martins W, Mathias J, Babenko D, Toleman MA, Engineering the Human Microbiome, A Natural Double-Barreled Approach Towards Solving both our Current Antibiotic Resistance and Sepsis Problems. Am J Biomed Sci & Res. 2020 - 11(3). AJBSR.MS.ID.001632. DOI: 10.34297/AJBSR.2020.11.001632. Received: December 12, 2020; Published: December 18, 2020 Opinion resistance mechanism responsible for this rise was an enzyme Many of the bacteria that cause life-threatening infection live called CTX-M-15 that gave resistance to the penicillin, monobactam, in very close association with us. Escherichia coli, for example and cephalosporin classes of the ß-lactam family of antibiotics (c. causes 80% of common community associated urinary tract infections and is also the main cause of serious (blood-stream) in several enteric pathogens including E. coli in New Delhi, India infection throughout Europe [1,2]. However, E. coli routinely lives 50% of all antibiotics) [7]. Its gene blaCTX-M-15 was first isolated in 2000 [8]. Later publications indicated that it was widespread as a commensal organism in our gut. -
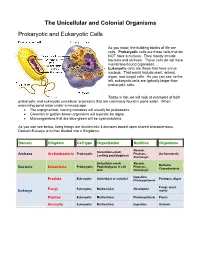
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

Structural Basis of Mammalian Mucin Processing by the Human Gut O
ARTICLE https://doi.org/10.1038/s41467-020-18696-y OPEN Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila ✉ ✉ Beatriz Trastoy 1,4, Andreas Naegeli2,4, Itxaso Anso 1,4, Jonathan Sjögren 2 & Marcelo E. Guerin 1,3 Akkermansia muciniphila is a mucin-degrading bacterium commonly found in the human gut that promotes a beneficial effect on health, likely based on the regulation of mucus thickness 1234567890():,; and gut barrier integrity, but also on the modulation of the immune system. In this work, we focus in OgpA from A. muciniphila,anO-glycopeptidase that exclusively hydrolyzes the peptide bond N-terminal to serine or threonine residues substituted with an O-glycan. We determine the high-resolution X-ray crystal structures of the unliganded form of OgpA, the complex with the glycodrosocin O-glycopeptide substrate and its product, providing a comprehensive set of snapshots of the enzyme along the catalytic cycle. In combination with O-glycopeptide chemistry, enzyme kinetics, and computational methods we unveil the molecular mechanism of O-glycan recognition and specificity for OgpA. The data also con- tribute to understanding how A. muciniphila processes mucins in the gut, as well as analysis of post-translational O-glycosylation events in proteins. 1 Structural Biology Unit, Center for Cooperative Research in Biosciences (CIC bioGUNE), Basque Research and Technology Alliance (BRTA), Bizkaia Technology Park, Building 801A, 48160 Derio, Spain. 2 Genovis AB, Box 790, 22007 Lund, Sweden. 3 IKERBASQUE, Basque Foundation for Science, 48013 ✉ Bilbao, Spain. 4These authors contributed equally: Beatriz Trastoy, Andreas Naegeli, Itxaso Anso. -

Infection Control
infection control JANICE CARR The above photo depicts an E.coli (ATCC 11775) biofilm grown on PC (polycarbonate) coupons using a CDC biofilm reactor. Microorganisms often colonize, and adhere strongly to living and non-living surfaces forming biofilms, and at times, demonstrate an increased resistance to antimicrobials. Biofilms on indwelling medical devices pose a serious threat to public health. BIOFILMS:BIOFILMS: Friend or Foe? By NICOLE KENNY, B.Sc, Assoc.Chem., Director of Professional & Technical Services, Virox Technologies Inc 38 Sanitation Canada - SEPTEMBER / OCTOBER 2006 JANICE CARR Scanning electron micrograph of a Staphylococcus biofilm on the inner surface of a needleless connector. A distinguishing characteristic of biofilms is the presence of extracellular polymeric substances, primarily polysaccharides, surrounding and encasing the cells. Here, there polysaccharides have been visualized by scanning electron microscopy. Picture yourself a con- “Why didn’t I listen to my mother and take Biofilms can be dangerous or benefi- testant on Jeopardy. Alex more science courses?” But in that split cial depending on where they are found Tribec has just asked you second you also remember a documen- and of which organisms they are com- to choose the category. tary you watched on CNN about whirl- prised. In industry, biofilms are responsi- You’re lagging behind the pool tubs and you know the answer. ble for billions of dollars in lost produc- leader by $400. All but “Alex, what are BIOFILMS?” tivity due to equipment damage, notori- one of the $500 questions ously famous for causing pipes to plug or Phave been taken, and the last category has THE ISSUE corrode. -

Current Issues of Nano-Bio-Science
Current Issues of Nano-Bio-Science CeNS Winterschool 2003 Mauterndorf, Austria 24-28 February 2003 Current Issues of Nano-Bio-Science CeNS Winterschool 2003 Mauterndorf, Austria 24-28 February 2003 in cooperation with SFB 486 and SFB 513 Program Committee Organisation Christoph Bräuchle Monika Kaempfe Jan von Delft Evelyn Morgenroth Hermann Gaub Joachim Rädler Jörg P. Kotthaus Paul Leiderer Joachim Rädler Internal Students’ Seminar: Schedule Saturday, 22nd February 7:30 pm Meeting in front of the castle’s lower gate to go the “Skialm” together 8:00 pm Get-together at the „Skialm“ in Mauterndorf (in the Skiing center) (kitchen closes at 9:30 pm) Sunday, 23rd February 9:45 am Introduction (W. Parak, F. Simmel) 10:00 am Simon Keller (AG Rädler) 10:30 am Stefan Griessl (AG Heckl) 11:00 am Coffee break 11:30 am Christine Meyer (AG Kotthaus) 12:00 pm Michael Sindel (AG von Delft) 12:30 pm Lunch (not provided), informal discussions 5:00 pm Ferdinand Kühner (AG Gaub) 5:30 pm Stefan Beyer (NG Simmel) Teresa Pellegrino (NG Parak) 6:00 pm Stefan Kowarik (AG Feldmann) 6:30 pm Break 7:00 pm Niklolay Petkov (AG Bein) 7:30 pm Sebastian Gritschneder (AG Reichling) 8:00 pm Ralf Bausinger / Johanna Kirstein (AG Bräuchle) 8:30 pm Conclusion Program Monday, 24th February 2003 8.30 – 8.45 Opening 8.45 – 9.45 Cees Dekker, Delft University of Technology Carbon nanotubes as model systems for nanoelectronics and nanosensors 9.45 – 10.45 Ulf Diederichsen, Universität Göttingen Molecular architecture with biooligomers 10.45 – 11.15 Coffee Break 11.15 – 12.15 Wolfgang -

Functional Anatomy of Prokaryotes and Eukaryotes
FUNCTIONAL ANATOMY OF PROKARYOTES AND EUKARYOTES BY DR JAWAD NAZIR ASSISTANT PROFESSOR DEPARTMENT OF MICROBIOLOGY UNIVERSITY OF VETERINARY AND ANIMAL SCIENCES, LAHORE Prokaryotes vs Eukaryotes Prokaryote comes from the Greek words for pre-nucleus Eukaryote comes from the Greek words for true nucleus. Functional anatomy of prokaryotes Prokaryotes vs Eukaryotes Prokaryotes Eukaryotes One circular chromosome, not in Paired chromosomes, in nuclear a membrane membrane No histones Histones No organelles Organelles Peptidoglycan cell walls Polysaccharide cell walls Binary fission Mitotic spindle Functional anatomy of prokaryotes Size and shape Average size: 0.2 -1.0 µm 2 - 8 µm Basic shapes: Functional anatomy of prokaryotes Size and shape Pairs: diplococci, diplobacilli Clusters: staphylococci Chains: streptococci, streptobacilli Functional anatomy of prokaryotes Size and shape Functional anatomy of prokaryotes Size and shape Functional anatomy of prokaryotes Size and shape Unusual shapes Star-shaped Stella Square Haloarcula Most bacteria are monomorphic A few are pleomorphic Genus: Stella Genus: Haloarcula Functional anatomy of prokaryotes Bacterial cell structure Structures external to cell wall Cell wall itself Structures internal to cell wall Functional anatomy of prokaryotes Glycocalyx Outside cell wall Usually sticky A capsule is neatly organized A slime layer is unorganized & loose Extracellular polysaccharide allows cell to attach Capsules prevent phagocytosis Association with diseases B. anthracis S. pneumoniae Functional anatomy of prokaryotes Flagella Outside cell wall Filament made of chains of flagellin Attached to a protein hook Anchored to the wall and membrane by the basal body Functional anatomy of prokaryotes Flagella Arrangement Functional anatomy of prokaryotes Bacterial motility Rotate flagella to run or tumble Move toward or away from stimuli (taxis) Flagella proteins are H antigens (e.g., E. -

Cell Structure and Function in the Bacteria and Archaea
4 Chapter Preview and Key Concepts 4.1 1.1 DiversityThe Beginnings among theof Microbiology Bacteria and Archaea 1.1. •The BacteriaThe are discovery classified of microorganismsinto several Cell Structure wasmajor dependent phyla. on observations made with 2. theThe microscope Archaea are currently classified into two 2. •major phyla.The emergence of experimental 4.2 Cellscience Shapes provided and Arrangements a means to test long held and Function beliefs and resolve controversies 3. Many bacterial cells have a rod, spherical, or 3. MicroInquiryspiral shape and1: Experimentation are organized into and a specific Scientificellular c arrangement. Inquiry in the Bacteria 4.31.2 AnMicroorganisms Overview to Bacterialand Disease and Transmission Archaeal 4.Cell • StructureEarly epidemiology studies suggested how diseases could be spread and 4. Bacterial and archaeal cells are organized at be controlled the cellular and molecular levels. 5. • Resistance to a disease can come and Archaea 4.4 External Cell Structures from exposure to and recovery from a mild 5.form Pili allowof (or cells a very to attach similar) to surfacesdisease or other cells. 1.3 The Classical Golden Age of Microbiology 6. Flagella provide motility. Our planet has always been in the “Age of Bacteria,” ever since the first 6. (1854-1914) 7. A glycocalyx protects against desiccation, fossils—bacteria of course—were entombed in rocks more than 3 billion 7. • The germ theory was based on the attaches cells to surfaces, and helps observations that different microorganisms years ago. On any possible, reasonable criterion, bacteria are—and always pathogens evade the immune system. have been—the dominant forms of life on Earth. -

What Are the Differences Between These Microbial Ecosystems? Wwwwe Now Know That, Like Other Org Gm,Anisms, Bacteria Exhibit Social Behaviors
What are the differences between these microbial ecosystems? WWwwe now know that, like other org gm,anisms, bacteria exhibit social behaviors. Bacterial cell escaping for a rare moment of peace and quiet contemplation Sociomicrobiology I. Cell signaling A. Definition/description B. Intraspecific (within spp.): Myxococcus C. Interspecific (between spp.): Pseudomonas aureofaciens II. Biofilms A. Biofilm formation B. Planktonic cells vs. biofilm cells C. General characteristics, structures D. Biofilms as social entities Small diffusible molecules mediate bacterial communication O O N H AHL O O O N H O http://www.ted.com/index.php/talks/bonnie_bassler_on_how_bacteria_communicate.html Why cell-cell signaling in bacteria? Often, single cells in a population might benefit from knowing how many cells are present… “A multitude of bacteria are stronger than a few, thus by union are able overcome obstacles too great for few.” -- Dr. Erwin F. Smith, 1905 (Father of Plant Bacteriology) Pseudomonas aeruginosa in lungs Xylella fastidiosa in xylem Why cell-cell signaling in bacteria? Cell-cell signaling enables bacteria to coordinate behavior to respond quickly to environmental stimuli… such as: -presence of suitable host -change in nutrient availability -defense/competition against other microorganisms -many others! CmmiCommunica tion among btbacteri i:a: The example of Myxococcus xanthus, the Wolf Pack feeder of bacteria http://cmgm.stanford.edu/devbio/kaiserlab/about_myxo/about_myxococcus.html Cooperation among cells in a population: myxobacteria. The myxobacteria are Gram-negative, ubiquitous, soil-dwelling bacteria that are capable of multicellular, social behaviour. In the presence of nutrients, “swarms” of myxobacteria feed cooperatively by sharing extracellular digestive enzymes, and can prey on other bacteria.