Killing of Gram-Negative Bacteria by Lactoferrin and Lysozyme
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prokaryotes (Domains Bacteria & Archaea)
2/4/15 Prokaryotes (Domains Bacteria & Archaea) KEY POINTS 1. Decomposers: recycle organic and inorganic molecules in environment; makes them available to other organisms. 2. Essential components of symbioses. 3. Encompasses the origins of metabolism and metabolic diversity. 4. Origin of photosynthesis and formation of atmospheric Oxygen Ceno- Meso- zoic zoic ANTIQUITY Humans Paleozoic Colonization of land Animals Origin of solar system and Earth • >3.5 BILLION years old. • Alone for 2 1 4 billion years Proterozoic Archaean Prokaryotes Billions of 2 years ago3 Multicellular eukaryotes Single-celled eukaryotes Atmospheric oxygen General characteristics 1. Small: compare to 10-100µm for 0.5-5µm eukaryotic cell; single-celled; may form colonies. 2. Lack membrane- enclosed organelles. 3. Cell wall present, but different from plant cell wall. 1 2/4/15 General characteristics 4. Occur everywhere, most numerous organisms. – More individuals in a handful of soil then there are people that have ever lived. – By far more individuals in our gut than eukaryotic cells that are actually us. General characteristics 5. Metabolic diversity established nutritional modes of eukaryotes. General characteristics 6. Important decomposers and recyclers 2 2/4/15 General characteristics 6. Important decomposers and recyclers • Form the basis of global nutrient cycles. General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • About 1/2 of all human diseases are caused by Bacteria General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • Extremely important in agriculture as well. Pierce’s disease is caused by Xylella fastidiosa, a Gamma Proteobacteria. It causes over $56 million in damage annually in California. That’s with $34 million spent to control it! = $90 million in California alone. -

A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection
Published online: 2020-04-12 THIEME 302 A Champion of Host Defense: A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection Martin J. Page, BSc (Hons)1 Etheresia Pretorius, PhD1 1 Department of Physiological Sciences, Stellenbosch University, Address for correspondence Etheresia Pretorius, PhD, Department of Stellenbosch, South Africa Physiological Sciences, Faculty of Science, Stellenbosch University, Private Bag X1 Matieland, Stellenbosch, 7602, South Africa Semin Thromb Hemost 2020;46:302–319. (e-mail: [email protected]). Abstract Thrombocytopenia is commonly associated with sepsis and infections, which in turn are characterized by a profound immune reaction to the invading pathogen. Platelets are one of the cellular entities that exert considerable immune, antibacterial, and antiviral actions, and are therefore active participants in the host response. Platelets are sensitive to surrounding inflammatory stimuli and contribute to the immune response by multiple mechanisms, including endowing the endothelium with a proinflammatory phenotype, enhancing and amplifying leukocyte recruitment and inflammation, promoting the effector functions of immune cells, and ensuring an optimal adaptive immune response. During infection, pathogens and their products influence the platelet response and can even be toxic. However, platelets are able to sense and engage bacteria and viruses to assist in their removal and destruction. Platelets greatly contribute to host defense by multiple mechanisms, including forming immune complexes and aggregates, shedding their granular content, and internalizing pathogens and subsequently being marked for removal. These processes, and the nature of platelet function in general, cause the platelet to be irreversibly consumed in Keywords the execution of its duty. An exaggerated systemic inflammatory response to infection ► platelets can drive platelet dysfunction, where platelets are inappropriately activated and face ► virus immunological destruction. -

Of Penicillin G, Other Penicillins, and a Cephalosporin
GLYCOPEPTIDE TRANSPEPTIDASE AND D-ALANINE CARBOXYPEPTIDASE: PENICILLIN-SENSITIVE ENZYMATIC REACTIONS* BY KAZUO IZAKI, M\ICHIO M\ATSUHASHI, AND JACK L. STROMINGER DEPARTMENT OF PHARMACOLOGY, UNIVERSITY OF WISCONSIN MEDICAL SCHOOL, MADISON Communicated by Clinton L. Woolsey, January 28, 1966 In 1929, Fleming discovered penicillins and observed that this group of substances kills gram-positive bacteria more effectively than gram-negative bacteria (thus im- plying some important physiological difference between the two groups of or- ganisms).1 Subsequent knowledge of the bacterial cell wall made it possible to establish both on physiological and chemical grounds that penicillins are specific and highly selective inhibitors of the biosynthesis of cell walls, both in gram-positive and in gram-negative bacteria.2-6 The reason for the relative insensitivity of gram- negative bacteria to most penicillins has remained obscure. More recent knowledge of the structure and biosynthesis of bacterial cell walls has suggested that, in a terminal reaction in cell wall synthesis, linear glycopeptide strands are cross-linked in a transpeptidation, accompanied by release of the termi- nal D-alanine of the pentapeptide precursor, with formation of a two- or three- dimensional network. Direct chemical analyses of cell walls prepared from cells treated with penicillin7' 8 and isotopic studies of wall biosynthesis9' 10 suggested that penicillin was interfering with this hypothetical glycopeptide cross-linking reaction. In the presence of penicillin, nascent glycopeptide, -
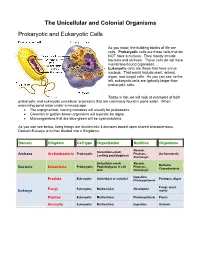
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

CARBAPENEMASE PRODUCING Enterobacteriaceae: MOLECULAR EPIDEMIOLOGY and ASSESSMENT of ALTERNATIVE THERAPEUTIC OPTIONS
CARBAPENEMASE PRODUCING Enterobacteriaceae: MOLECULAR EPIDEMIOLOGY AND ASSESSMENT OF ALTERNATIVE THERAPEUTIC OPTIONS By LAURA JIMENA ROJAS COY Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Microbiology and Molecular Biology CASE WESTERN RESERVE UNIVERSITY May, 2018 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the dissertation of Laura Jimena Rojas Coy candidate for the degree of Doctor of Philosophy*. Committee Chair Dr. Arne Riestch Committee Member Dr. Liem Nguyen Committee Member Dr. Anthony Wynshaw-Boris Committee Member Dr. Robert A. Bonomo Date of Defense March 22, 2018 *We also certify that written approval has been obtained for any proprietary material contained therein. DEDICATION To my mother Maria Grelly and my father Daniel for raising me believing that with hard work I could achieve anything I set my mind to. To my loving brother Cami for constantly looking up to me, inspiring me to always do my best; and finally to my adorable little sister Angie for her infinite an unconditional love. I love you all so much. iii ACKNOWLEDGEMENTS My deepest gratitude and appreciation to my mentor, Dr. Robert A. Bonomo, for being so generous, understanding and patient and for giving me so many wonderful opportunities, in spite of not following the "traditional" path. In addition to the tremendous academic support, he has shown me by his example that humbleness, fairness and leadership is what truly makes a good scientist. To my committee members thank you for your valuable suggestions and criticisms. To everyone in the Bonomo Lab, thank you for sharing your expertise with me and, for constantly helping me, and for each and every one of your invaluable contributions to the work summarized in this dissertation. -

Structural Basis of Mammalian Mucin Processing by the Human Gut O
ARTICLE https://doi.org/10.1038/s41467-020-18696-y OPEN Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila ✉ ✉ Beatriz Trastoy 1,4, Andreas Naegeli2,4, Itxaso Anso 1,4, Jonathan Sjögren 2 & Marcelo E. Guerin 1,3 Akkermansia muciniphila is a mucin-degrading bacterium commonly found in the human gut that promotes a beneficial effect on health, likely based on the regulation of mucus thickness 1234567890():,; and gut barrier integrity, but also on the modulation of the immune system. In this work, we focus in OgpA from A. muciniphila,anO-glycopeptidase that exclusively hydrolyzes the peptide bond N-terminal to serine or threonine residues substituted with an O-glycan. We determine the high-resolution X-ray crystal structures of the unliganded form of OgpA, the complex with the glycodrosocin O-glycopeptide substrate and its product, providing a comprehensive set of snapshots of the enzyme along the catalytic cycle. In combination with O-glycopeptide chemistry, enzyme kinetics, and computational methods we unveil the molecular mechanism of O-glycan recognition and specificity for OgpA. The data also con- tribute to understanding how A. muciniphila processes mucins in the gut, as well as analysis of post-translational O-glycosylation events in proteins. 1 Structural Biology Unit, Center for Cooperative Research in Biosciences (CIC bioGUNE), Basque Research and Technology Alliance (BRTA), Bizkaia Technology Park, Building 801A, 48160 Derio, Spain. 2 Genovis AB, Box 790, 22007 Lund, Sweden. 3 IKERBASQUE, Basque Foundation for Science, 48013 ✉ Bilbao, Spain. 4These authors contributed equally: Beatriz Trastoy, Andreas Naegeli, Itxaso Anso. -

Functional Anatomy of Prokaryotes and Eukaryotes
FUNCTIONAL ANATOMY OF PROKARYOTES AND EUKARYOTES BY DR JAWAD NAZIR ASSISTANT PROFESSOR DEPARTMENT OF MICROBIOLOGY UNIVERSITY OF VETERINARY AND ANIMAL SCIENCES, LAHORE Prokaryotes vs Eukaryotes Prokaryote comes from the Greek words for pre-nucleus Eukaryote comes from the Greek words for true nucleus. Functional anatomy of prokaryotes Prokaryotes vs Eukaryotes Prokaryotes Eukaryotes One circular chromosome, not in Paired chromosomes, in nuclear a membrane membrane No histones Histones No organelles Organelles Peptidoglycan cell walls Polysaccharide cell walls Binary fission Mitotic spindle Functional anatomy of prokaryotes Size and shape Average size: 0.2 -1.0 µm 2 - 8 µm Basic shapes: Functional anatomy of prokaryotes Size and shape Pairs: diplococci, diplobacilli Clusters: staphylococci Chains: streptococci, streptobacilli Functional anatomy of prokaryotes Size and shape Functional anatomy of prokaryotes Size and shape Functional anatomy of prokaryotes Size and shape Unusual shapes Star-shaped Stella Square Haloarcula Most bacteria are monomorphic A few are pleomorphic Genus: Stella Genus: Haloarcula Functional anatomy of prokaryotes Bacterial cell structure Structures external to cell wall Cell wall itself Structures internal to cell wall Functional anatomy of prokaryotes Glycocalyx Outside cell wall Usually sticky A capsule is neatly organized A slime layer is unorganized & loose Extracellular polysaccharide allows cell to attach Capsules prevent phagocytosis Association with diseases B. anthracis S. pneumoniae Functional anatomy of prokaryotes Flagella Outside cell wall Filament made of chains of flagellin Attached to a protein hook Anchored to the wall and membrane by the basal body Functional anatomy of prokaryotes Flagella Arrangement Functional anatomy of prokaryotes Bacterial motility Rotate flagella to run or tumble Move toward or away from stimuli (taxis) Flagella proteins are H antigens (e.g., E. -

Protracted Anemia Associated with Chronic, Relapsing Systemic Inflammation Induced by Arthropathic Peptidoglycan-Polysaccharide Polymers in Rats R
INFECTION AND IMMUNITY, Apr. 1989, p. 1177-1185 Vol. 57, No. 4 0019-9567/89/041177-09$02.00/0 Copyright © 1989, American Society for Microbiology Protracted Anemia Associated with Chronic, Relapsing Systemic Inflammation Induced by Arthropathic Peptidoglycan-Polysaccharide Polymers in Rats R. BALFOUR SARTOR,l.2* SONIA K. ANDERLE,3 NADER RIFAI,4 DAVID A. T. GOO,4 WILLIAM J. CROMARTIE,3 AND JOHN H. SCHWAB2'3 Department of Medicine,1 Department of Microbiology and Immunology,3 Department ofPathology,4 and Core Center for Diarrheal Disease,2 University of North Carolina, Chapel Hill, North Carolina 27599-7080 Received 3 October 1988/Accepted 6 January 1989 Mild hypoproliferative anemia with abnormal iron metabolism frequently accompanies chronic inflamma- tion and infection in humans. To determine whether anemia is associated with chronic relapsing arthritis induced by bacterial cell wall polymers, serial determinations of the hematocrit were measured in rats injected intraperitoneally with sonicated peptidoglycan-polysaccharide fragments from group A streptococci. Acute anemia peaked 5 to 10 days after injection, and chronic, spontaneously relapsing anemia persisted for 309 days. 51Cr labeling demonstrated decreased erythrocyte survival, i.e., a half-life of 8.4 days in cell wall-injected rats versus 11.8 days in controls. Erythrocytes were mildly microcytic, and leukocyte counts were elevated during early spontaneous reactivation of arthritis, 15 days after injection of peptidoglycan-polysaccharide. Bone marrow myeloid/erythroid precursor ratios were elevated in arthritic rats (P < 0.0001). Purified peptidoglycan produced an acute anemia lasting 10 days, while injection of group A streptococcal polysaccharide and mutanolysin-digested cell wall did not affect the hematocrit. -

SUPPLEMENTARY MATERIAL 1 /FODOR ET AL Zoonic and Veterinary Pathogen Candidates for the “ESCAPE Club” S1.1 Escherichia Coli Commensal Strains of E
SUPPLEMENTARY MATERIAL 1 /FODOR ET AL Zoonic and veterinary pathogen candidates for the “ESCAPE Club” S1.1 Escherichia coli Commensal strains of E. coli, as versatile residents of the lower intestine, are also repeatedly challenged by antimicrobial pressures during the lifetime of their host. Consequently, commensal strains acquire resistance genes, and/or develop resistant mutants in order to survive and maintain microbial homeostasis in the lower intestinal tract. Commensal E. coli strains are regarded as indicators of the antimicrobial load of their hosts. The recent review (Szmolka, A. and Nagy, B. 2013) described the historical background of the origin, appearance and transfer mechanisms of antimicrobial resistance genes into an original animal - commensal intestinal E. coli with comparative information on their pathogenic counterparts. The most efficient mechanism used by E. coli against different antimicrobial-based efflux pumps and mobile resistance mechanisms carried by plasmids and/or mobile genetic elements is known. For a while, these mechanisms cannot protect E. coli against fabclavine (Fodor L. et al., in preparation). The emergence of hybrid plasmids, both resistance and virulent, among E. coli is of additional public concern. Co-existence and co-transfer of these “bad genes” in this huge and most versatile in vivo compartment may represent increased public health risk in the future. The significance of MDR commensal E. coli seems to be highest in the food animal industry, which may function as a reservoir for intra- and interspecific exchange, and a source for the spread of MDR determinants through contaminated food to humans. Thus, the potential of MDR occurring in these commensal bacteria living in animals used as sources of food (as meat, eggs, milk) should be a concern from the aspect of public health, and it needs to be continuously monitored in the future by using the toolkit of molecular genetics [8]. -

Evolution of Vancomycin-Resistant Enterococcus Faecium During Colonization and Infection in Immunocompromised Pediatric Patients
Evolution of vancomycin-resistant Enterococcus faecium during colonization and infection in immunocompromised pediatric patients Gayatri Shankar Chilambia, Hayley R. Nordstroma, Daniel R. Evansa, Jose A. Ferrolinob, Randall T. Haydenc, Gabriela M. Marónb,d, Anh N. Vob,d, Michael S. Gilmoree,f, Joshua Wolfb,d,1, Jason W. Roschb,1, and Daria Van Tynea,1,2 aDivision of Infectious Diseases, University of Pittsburgh School of Medicine, Pittsburgh, PA 15213; bDepartment of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, TN 38105; cDepartment of Pathology, St. Jude Children’s Research Hospital, Memphis, TN 38105; dDepartment of Pediatrics, University of Tennessee Health Science Center, Memphis, TN 38105; eDepartment of Ophthalmology, Harvard Medical School and Massachusetts Eye and Ear Infirmary, Boston, MA 02114; and fDepartment of Microbiology, Harvard Medical School, Boston, MA 02115 Edited by Paul E. Turner, Yale University, New Haven, CT, and approved April 8, 2020 (received for review October 13, 2019) Patients with hematological malignancies or undergoing hemato- in the hospital environment poses a major threat to patient poietic stem cell transplantation are vulnerable to colonization safety, and vancomycin-resistant E. faecium (VREfm) carrying and infection with multidrug-resistant organisms, including VanA-type resistance currently pose the biggest threat in the vancomycin-resistant Enterococcus faecium (VREfm). Over a 10-y United States (10–12). period, we collected and sequenced the genomes of 110 VREfm Immunocompromised patients are prone to drug-resistant isolates from gastrointestinal and blood cultures of 24 pediatric enterococcal infection, due to frequent use of broad-spectrum patients undergoing chemotherapy or hematopoietic stem cell antibiotics in hospital environments where resistant lineages transplantation for hematological malignancy at St. -

Overcoming Inhibitor Resistance in the Shv Βeta
OVERCOMING INHIBITOR RESISTANCE IN THE SHV ΒETA-LACTAMASE By JODI MICHELLE THOMSON Submitted in partial fulfillment of the requirements For the degree of Doctor of Philosophy Dissertation Advisor: Robert A Bonomo Department of Pharmacology CASE WESTERN RESERVE UNIVERSITY August, 2007 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the dissertation of ______________________________________________________ candidate for the Ph.D. degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. For Robert A. Bonomo TABLE OF CONTENTS Table of Contents ............................................................................... 1-2 List of Figures ..................................................................................... 3-8 List of Tables...........................................................................................9 Acknowledgments........................................................................... 10-12 Abbreviations .................................................................................. 13-14 Abstract.......................................................................................... -

Elucidating Peptidoglycan Structure: an Analytical Toolset
Trends in Microbiology Review Elucidating Peptidoglycan Structure: An Analytical Toolset Sara Porfírio,1 Russell W. Carlson,1 and Parastoo Azadi1,* Peptidoglycan (PG) is a ubiquitous structural polysaccharide of the bacterial cell Highlights wall, essential in preserving cell integrity by withstanding turgor pressure. Any PG is a major bacterial cell wall compo- change that affects its biosynthesis or degradation will disturb cell viability, nent, crucial for cell integrity, and strongly therefore PG is one of the main targets of antimicrobial drugs. Considering its related to antibiotic sensitivity. major role in cell structure and integrity, the study of PG is of utmost relevance, Methods to analyze PG structure have with prospective ramifications to several disciplines such as microbiology, phar- evolved over time, and new and im- macology, agriculture, and pathogenesis. Traditionally, high-performance liquid proved methods are currently replacing chromatography (HPLC) has been the workhorse of PG analysis. In recent years, previous tedious and long procedures. technological and bioinformatic developments have upgraded this seminal Current tools, ranging from technique, making analysis more sensitive and efficient than ever before. Here chromatography–mass spectrometry we describe a set of analytical tools for the study of PG structure (from composi- methods to imaging techniques, allow tion to 3D architecture), identify the most recent trends, and discuss future characterizing PG from a structural and architectural perspective. challenges in the field. Peptidoglycan Structure and Diversity Peptidoglycan (PG) (also known as murein, from Latin ‘murus’, wall) is a major component of the bacterial cell wall [1] (Figure 1). Present in both Gram-positive and Gram-negative bacteria [2], this polysaccharide forms a flexible, net-like structure that determines cell shape and provides mechanical strength and osmotic stability.