Venomics and Cellular Toxicity of Thai Pit Vipers (Trimeresurus Macrops and T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Working with a Full Deck: the Use of Picture Cards in Herpetological Surveys of Timor-Leste
68 TECHNIQUES LITERATURE CITED NOUVELLET, P., G. S. A. RASMUSSEN, D. W. MACDONALD, AND F. COURCHAMP. 2012. Noisy clocks and silent sunrises: measurement methods of BURGER, B. 1988. Which way did he go? Telonics Quarterly 1:1. daily activity patterns. J. Zool., London 286:179–184. DORCAS, M. E., AND C. R. PETERSON. 2012. Automated data acquisi- RODDA, G. H. 1984. Movements of juvenile American crocodiles in Ga- tion. In R. W. McDiarmid, M. S. Foster, C. Guyer, J. Gibbons, and tun Lake, Panama. Herpetologica 40:444–451. N. Chernoff (eds.), Reptile Biodiversity: Standard Methods for In- ventory and Monitoring, pp. 61–68. University of California Press, Berkeley, California. Herpetological Review, 2013, 44(1), 68–76. © 2013 by Society for the Study of Amphibians and Reptiles Working with a Full Deck: the Use of Picture Cards in Herpetological Surveys of Timor-Leste Timor is the 44th largest island in the world and the seventh significantly, the Muséum National d’Histoire Naturelle in Paris, largest between Asia and Australia (area 29,402 km2). It occupies France; Naturalis, formerly the Rijksmuseum van Natuurlijke an extremely interesting geographical position within the bio- Historie, in Leiden, The Netherlands; and the Zoologisch Mu- geographical sub-region known as Wallacea, at the southeastern seum Amsterdam, now also housed in Leiden). Additional short edge of the Lesser Sunda Archipelago and separated from Aus- surveys were conducted there during the early 20th century (e.g., tralia by the Timor Sea (ca. 450 km). This gap was considerably Smith 1927; collections in the Natural History Museum, London, lessened during the final 250,000 years of the Pleistocene Epoch United Kingdom) and in the 1990s (e.g., How et al. -

Epidemiology of Snakebites from a General Hospital in Singapore: a 5-Year Retrospective Review (2004-2008) 1 Hock Heng Tan, MBBS, FRCS A&E (Edin), FAMS
640 Epidemiology of Snakebites—Hock Heng Tan Original Article Epidemiology of Snakebites from A General Hospital in Singapore: A 5-year Retrospective Review (2004-2008) 1 Hock Heng Tan, MBBS, FRCS A&E (Edin), FAMS Abstract Introduction: This is a retrospective study on the epidemiology of snakebites that were presented to an emergency department (ED) between 2004 and 2008. Materials and Methods: Snakebite cases were identified from International Classification of Diseases (ICD) code E905 and E906, as well as cases referred for eye injury from snake spit and records of antivenom use. Results: Fifty-two cases were identified: 13 patients witnessed the snake biting or spitting at them, 22 patients had fang marks and/or clinical features of envenomations and a snake was seen and the remaining 17 patients did not see any snake but had fang marks suggestive of snakebite. Most of the patients were young (mean age 33) and male (83%). The three most commonly identified snakes were cobras (7), pythons (4) and vipers (3). One third of cases occurred during work. Half of the bites were on the upper limbs and about half were on the lower limbs. One patient was spat in the eye by a cobra. Most of the patients (83%) arrived at the ED within 4 hours of the bite. Pain and swelling were the most common presentations. There were no significant systemic effects reported. Two patients had infection and 5 patients had elevated creatine kinase (>600U/L). Two thirds of the patients were admitted. One patient received antivenom therapy and 5 patients had some form of surgical intervention, of which 2 had residual disability. -

Multi-National Conservation of Alligator Lizards
MULTI-NATIONAL CONSERVATION OF ALLIGATOR LIZARDS: APPLIED SOCIOECOLOGICAL LESSONS FROM A FLAGSHIP GROUP by ADAM G. CLAUSE (Under the Direction of John Maerz) ABSTRACT The Anthropocene is defined by unprecedented human influence on the biosphere. Integrative conservation recognizes this inextricable coupling of human and natural systems, and mobilizes multiple epistemologies to seek equitable, enduring solutions to complex socioecological issues. Although a central motivation of global conservation practice is to protect at-risk species, such organisms may be the subject of competing social perspectives that can impede robust interventions. Furthermore, imperiled species are often chronically understudied, which prevents the immediate application of data-driven quantitative modeling approaches in conservation decision making. Instead, real-world management goals are regularly prioritized on the basis of expert opinion. Here, I explore how an organismal natural history perspective, when grounded in a critique of established human judgements, can help resolve socioecological conflicts and contextualize perceived threats related to threatened species conservation and policy development. To achieve this, I leverage a multi-national system anchored by a diverse, enigmatic, and often endangered New World clade: alligator lizards. Using a threat analysis and status assessment, I show that one recent petition to list a California alligator lizard, Elgaria panamintina, under the US Endangered Species Act often contradicts the best available science. -

Final Report for the University of Nottingham / Operation Wallacea Forest Projects, Honduras 2004
FINAL REPORT for the University of Nottingham / Operation Wallacea forest projects, Honduras 2004 TABLE OF CONTENTS FINAL REPORT FOR THE UNIVERSITY OF NOTTINGHAM / OPERATION WALLACEA FOREST PROJECTS, HONDURAS 2004 .....................................................................................................................................................1 INTRODUCTION AND OVERVIEW ..............................................................................................................................3 List of the projects undertaken in 2004, with scientists’ names .........................................................................4 Forest structure and composition ..................................................................................................................................... 4 Bat diversity and abundance ............................................................................................................................................ 4 Bird diversity, abundance and ecology ............................................................................................................................ 4 Herpetofaunal diversity, abundance and ecology............................................................................................................. 4 Invertebrate diversity, abundance and ecology ................................................................................................................ 4 Primate behaviour........................................................................................................................................................... -

WHO Guidance on Management of Snakebites
GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition 1. 2. 3. 4. ISBN 978-92-9022- © World Health Organization 2016 2nd Edition All rights reserved. Requests for publications, or for permission to reproduce or translate WHO publications, whether for sale or for noncommercial distribution, can be obtained from Publishing and Sales, World Health Organization, Regional Office for South-East Asia, Indraprastha Estate, Mahatma Gandhi Marg, New Delhi-110 002, India (fax: +91-11-23370197; e-mail: publications@ searo.who.int). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by the World Health Organization to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either expressed or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. -

Trimeresurus Sp
Trimeresurus sp. Copyright: Auszug aus Datenbank der Toxikologischen Abteilung der II. Medizinischen Klinik München; Toxinfo von Kleber JJ , Ganzert M, Zilker Th; Ausgabe 2002; erstellt Wagner Ph, Kleber JJ; Korthals Altes 1999 TOXIKOLOGIE: bei allen Arten von Trimeresurus Sp kommt es immer zu Lokalsymptomen bis Nekrose; zu rechnen ist mit außerdem mit Gerinnungsstörungen, Schocksymptomen T. ALBOLABRIS: massive Lokalsymptome, Gerinnungsstörung leicht bei 30%, stark bei 10% (16); Letalität in Thailand 3% (12) T. FLAVOVIRIDIS: vor Antiserum-Zeit 15% Letalität (12) starke Schwellung + Nekrosen, Schock, keine Gerinnungsstörungen bisher berichtet (11,12) T. GRAMINEUS: Schwellung; keine Nekrosen, keine Gerinnungsstörungen berichtet (1,14) T. KANBURIENSIS: Schwellung, Schock, Gerinnungsstörung (12) T. MUCROSQUAMATUS: Schwellung, Gerinnungsstörungen (12) T. POPEIORUM: Lokalsymptome; sehr geringe Gerinnungsstörung mit normalem Fibrinogen + Thrombo (15) T. PURPUREOMACULATUS: Schwellung, Nekrose, Gerinnungsstörung bis 40% (12,15) T. WAGLERI: Schwellung, Gerinnungsstörung (15) SYMPTOME: erste Vergiftungssymptome direkt nach dem Biß (sofortiger Schmerz, Schwellung entwickelt sich in den ersten 2-4 h) (2); meist starke Schwellung (häufig halbes bis ganzes Glied), bis ca. eine Woche anhaltend; Lymphangitis und schmerzhafte Lymphknotenschwellung (1,2,5,6) ; lokale subkutane Hämorrhagie, gelegentlich Blasenbildung und Hautnekrosen (1,2); bei T. flavoviridis auch Muskelnekrosen und Kompartmentsyndrom (3,12) MUND: lokal nach Giftaussaugen Schwellung an Lippe + Zunge bei T. albolabris (12) COR: selten Butdruckabfall, Schock (11,12); selten EKG-Veränderungen bei T. mucrosquamatus (12) LABOR: Thrombin ähnliche Aktivität führt zur Defibrinierung bis Verbrauchskoagulopathie mit Hypo- bis Afibrinogenämie, Thrombopenie (auch erst nach 12h Latenz) (1,2,4,13,16); Aktivierung der Fibrinolyse mit später Plasminerniedrigung (16); Leukozytose SONST: häufig Übelkeit, Erbrechen, Bauchschmerzen (11, 12); selten Nierenschädigung berichtet bei T. -

On Trimeresurus Sumatranus
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/266262458 On Trimeresurus sumatranus (Raffles, 1822), with the designation of a neotype and the description of a new species of pitviper from Sumatra (Squamata: Viperidae: Crotalinae) Article in Amphibian and Reptile Conservation · September 2014 CITATIONS READS 4 360 3 authors, including: Gernot Vogel Irvan Sidik Independent Researcher Indonesian Institute of Sciences 102 PUBLICATIONS 1,139 CITATIONS 12 PUBLICATIONS 15 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Save Vietnam Biodiversity View project Systematics of the genus Pareas View project All content following this page was uploaded by Gernot Vogel on 01 October 2014. The user has requested enhancement of the downloaded file. Comparative dorsal view of the head of Trimeresurus gunaleni spec. nov. (left) and T. sumatranus (right). Left from above: male, female (holotype), male, all alive, from Sumatra Utara Province, Sumatra. Right: adult female alive from Bengkulu Province, Su- matra, adult male alive from Bengkulu Province, Sumatra, preserved female from Borneo. Photos: N. Maury. Amphib. Reptile Conserv. | amphibian-reptile-conservation.org (1) September 2014 | Volume 8 | Number 2 | e80 Copyright: © 2014 Vogel et al. This is an open-access article distributed under the terms of the Creative Commons Attribution–NonCommercial–NoDerivs 3.0 Unported License, Amphibian & Reptile Conservation which permits -

2019 Fry Trimeresurus Genus.Pdf
Toxicology Letters 316 (2019) 35–48 Contents lists available at ScienceDirect Toxicology Letters journal homepage: www.elsevier.com/locate/toxlet Clinical implications of differential antivenom efficacy in neutralising coagulotoxicity produced by venoms from species within the arboreal T viperid snake genus Trimeresurus ⁎ Jordan Debonoa, Mettine H.A. Bosb, Nathaniel Frankc, Bryan Frya, a Venom Evolution Lab, School of Biological Sciences, University of Queensland, St Lucia, QLD, 4072, Australia b Division of Thrombosis and Hemostasis, Einthoven Laboratory for Vascular and Regenerative Medicine, Leiden University Medical Center, Albinusdreef 2, 2333 ZA, Leiden, the Netherlands c Mtoxins, 1111 Washington Ave, Oshkosh, WI, 54901, USA ARTICLE INFO ABSTRACT Keywords: Snake envenomation globally is attributed to an ever-increasing human population encroaching into snake Venom territories. Responsible for many bites in Asia is the widespread genus Trimeresurus. While bites lead to hae- Coagulopathy morrhage, only a few species have had their venoms examined in detail. We found that Trimeresurus venom Fibrinogen causes haemorrhaging by cleaving fibrinogen in a pseudo-procoagulation manner to produce weak, unstable, Antivenom short-lived fibrin clots ultimately resulting in an overall anticoagulant effect due to fibrinogen depletion. The Phylogeny monovalent antivenom ‘Thai Red Cross Green Pit Viper antivenin’, varied in efficacy ranging from excellent neutralisation of T. albolabris venom through to T. gumprechti and T. mcgregori being poorly neutralised and T. hageni being unrecognised by the antivenom. While the results showing excellent neutralisation of some non-T. albolabris venoms (such as T. flavomaculaturs, T. fucatus, and T. macrops) needs to be confirmed with in vivo tests, conversely the antivenom failure T. -
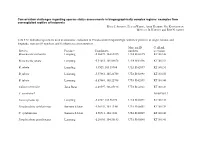
Conservation Challenges Regarding Species Status Assessments in Biogeographically Complex Regions: Examples from Overexploited Reptiles of Indonesia KYLE J
Conservation challenges regarding species status assessments in biogeographically complex regions: examples from overexploited reptiles of Indonesia KYLE J. SHANEY, ELIJAH WOSTL, AMIR HAMIDY, NIA KURNIAWAN MICHAEL B. HARVEY and ERIC N. SMITH TABLE S1 Individual specimens used in taxonomic evaluation of Pseudocalotes tympanistriga, with their province of origin, latitude and longitude, museum ID numbers, and GenBank accession numbers. Museum ID GenBank Species Province Coordinates numbers accession Bronchocela cristatella Lampung -5.36079, 104.63215 UTA R 62895 KT180148 Bronchocela jubata Lampung -5.54653, 105.04678 UTA R 62896 KT180152 B. jubata Lampung -5.5525, 105.18384 UTA R 62897 KT180151 B. jubata Lampung -5.57861, 105.22708 UTA R 62898 KT180150 B. jubata Lampung -5.57861, 105.22708 UTA R 62899 KT180146 Calotes versicolor Jawa Barat -6.49597, 106.85198 UTA R 62861 KT180149 C. versicolor* NC009683.1 Gonocephalus sp. Lampung -5.2787, 104.56198 UTA R 60571 KT180144 Pseudocalotes cybelidermus Sumatra Selatan -4.90149, 104.13401 UTA R 60551 KT180139 P. cybelidermus Sumatra Selatan -4.90711, 104.1348 UTA R 60549 KT180140 Pseudocalotes guttalineatus Lampung -5.28105, 104.56183 UTA R 60540 KT180141 P. guttalineatus Sumatra Selatan -4.90681, 104.13457 UTA R 60501 KT180142 Pseudocalotes rhammanotus Lampung -4.9394, 103.85292 MZB 10804 KT180147 Pseudocalotes species 4 Sumatra Barat -2.04294, 101.31129 MZB 13295 KT211019 Pseudocalotes tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60544 KT180143 P. tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60547 KT180145 Pogona vitticeps* AB166795.1 *Entry to GenBank by previous authors TABLE S2 Reptile species currently believed to occur Java and Sumatra, Indonesia, with IUCN Red List status, and certainty of occurrence. -

Reproductive Biology and Natural History of the White-Lipped Pit Viper (Trimeresurus Albolabris Gray, 1842) in Hong Kong Anne Devan-Song University of Rhode Island
University of Rhode Island DigitalCommons@URI Natural Resources Science Faculty Publications Natural Resources Science 2017 Reproductive Biology and Natural History of the White-lipped Pit Viper (Trimeresurus albolabris Gray, 1842) in Hong Kong Anne Devan-Song University of Rhode Island Paolo Martelli See next page for additional authors Follow this and additional works at: https://digitalcommons.uri.edu/nrs_facpubs Citation/Publisher Attribution Devan-Song, A., Martelli, P., & Karraker, N. E. (2017). Reproductive Biology and Natural History of the White-lipped Pit Viper (Trimeresurus albolabris Gray, 1842) in Hong Kong. Herpetological Conservation and Biology, 12(1), 41-55. Retrieved from http://www.herpconbio.org/Volume_12/Issue_1/Devan-Song_etal_2017.pdf Available at: http://www.herpconbio.org/Volume_12/Issue_1/Devan-Song_etal_2017.pdf This Article is brought to you for free and open access by the Natural Resources Science at DigitalCommons@URI. It has been accepted for inclusion in Natural Resources Science Faculty Publications by an authorized administrator of DigitalCommons@URI. For more information, please contact [email protected]. Authors Anne Devan-Song, Paolo Martelli, and Nancy E. Karraker This article is available at DigitalCommons@URI: https://digitalcommons.uri.edu/nrs_facpubs/115 Herpetological Conservation and Biology 12:41–55. Submitted: 30 September 2015; Accepted: 18 January 2017; Published: 30 April 2017. Reproductive Biology and Natural History of the White-lipped Pit Viper (Trimeresurus albolabris Gray, -

REPTILIA: SQUAMATA: CROTALIDAE Cerrophidion Tzotzilorum
880.1 REPTILIA: SQUAMATA: CROTALIDAE Cerrophidion tzotzilorum Catalogue of American Amphibians and Reptiles. Jadin, R.C. 2010. Cerrophidion tzotzilorum. Cerrophidion tzotzilorum (Campbell) Tzotzil Montane Pitviper Nauyaca del frío, víbora Bothrops nummifer mexicanus: McCoy and Van Horn 1962:186 (part). Cerrophidion godmani: Auth, Smith, Brown, and Lintz 2000:73 (part). _ FIGURE 2. Photograph of the hemipenis of the holotype Bothrops tzotzilorum Campbell 1985:48. Type locali- _ ty, “10.9 km ESE San Cristóbal de Las Casas, of Cerrophidion tzotzilorum (UTA R 9641); (A) sulcate view, Chiapas, Mexico, elevation 2320 m.” Holotype, (B) asulcate view. Amphibian and Reptile Diversity Research Cen- ter, University of Texas at Arlington (UTA) R_9641, • DIAGNOSIS. This species is parapatric with C. an adult male collected by J.A. Campbell on 8 godmani with few reports of sympatry (Campbell June 1979. 1985). It differs from C. godmani in having shorter Porthidium tzotzilorum: Campbell and Lamar 1989: and more numerous dorsal and lateral body blotches, 264. generally lower ventral scale counts, and fewer den- Cerrophidion tzotzilorum: Campbell and Lamar 1992: tary and pterygoid teeth. 24. • DESCRIPTIONS. The most complete descrip- • CONTENT. No subspecies are recognized. tions of the external morphology are in Campbell (1985, 1988), Campbell and Lamar (1989, 2004), and • DEFINITION. Cerrophidion tzotzilorum may be the Jadin (in press). smallest species of pitviper in the New World (Camp- bell and Lamar 2004); adults probably do not exceed • ILLUSTRATIONS. In the original description, 50 cm in total length. Coloration may be a dark gray- Campbell (1985) provided sketches of both the left ish brown or rust. A darker gray or brown zig_zag pat- side and dorsal views of the head of the holotype, tern extends from the neck down the entire length of along with a sulcate view of the left hemipene. -

Morphological and Genetic Verification of Ovophis Tonkinensis (Bourret, 1934) in Hong Kong
Herpetology Notes, volume 10: 457-461 (2017) (published online on 06 September 2017) Morphological and genetic verification of Ovophis tonkinensis (Bourret, 1934) in Hong Kong Jonathan J. Fong1, Alessandro Grioni2, Paul Crow2 and Ka-shing Cheung3,* Abstract. Hong Kong lies in the putative ranges of two Ovophis species: O. makazayazaya (Sichuan and Yunnan Provinces, Taiwan, and northern Vietnam) and O. tonkinensis (Guangxi, Guangdong, and Hainan Provinces, and northern Vietnam). It is unclear which Ovophis species is/are present in Hong Kong. Previous studies identified O. tonkinensis using morphology, but without molecular data. In this study, we verified the presence of O. tonkinensis in Hong Kong using molecular (four mitochondrial DNA loci) and morphological data of two specimens. The clade that is currently identified as Ovophis tonkinensis has a large geographic range and relatively deep divergences, hinting that its taxonomy requires further work. Key words. Hong Kong, Ovophis tonkinensis, Ovophis makazayazaya, reptile, snake. Introduction makazayazaya (Karsen et al. 1998). Malhotra et al. (2011) included four Ovophis specimens from Hong Kong The Asian pitviper genus Ovophis has had a (BMNH 1983 281–284) in their morphometric analyses. complicated taxonomic history. Previously, Ovophis These four specimens grouped with O. tonkinensis was believed to contain three species (O. chaseni specimens from northern Vietnam and southern China. (Smith, 1931), O. okinavensis (Boulenger, 1892), O. Studies in areas close to Hong Kong also confirmed the monticola (Gunther, 1864; 4 subspecies)) (McDiarmid presence of O. tonkinensis, including Hainan Province et al. 1999). However, Malhotra and Thorpe (2004) (morphology: David (1995), David (2001), Wang et removed the first two species from this genus so that al.