Kaposi Sarcoma Herpesvirus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Novel 2-Herpesvirus of the Rhadinovirus 2 Lineage In
Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Letter A Novel ␥2-Herpesvirus of the Rhadinovirus 2 Lineage in Chimpanzees Vincent Lacoste,1 Philippe Mauclère,1,2 Guy Dubreuil,3 John Lewis,4 Marie-Claude Georges-Courbot,3 andAntoine Gessain 1,5 1Unite´d’Epide´miologie et Physiopathologie des Virus Oncoge`nes, De´partement du SIDA et des Re´trovirus, Institut Pasteur, 75724 Paris Cedex 15, France; 2Centre Pasteur du Cameroun, BP 1274, Yaounde´, Cameroon; 3Centre International de Recherches Me´dicales, Franceville, Gabon; 4International Zoo Veterinary Group, Keighley, West Yorkshire BD21 1AG, UK Old World monkeys and, recently, African great apes have been shown, by serology and polymerase chain reaction (PCR), to harbor different ␥2-herpesviruses closely related to Kaposi’s sarcoma-associated Herpesvirus (KSHV). Although the presence of two distinct lineages of KSHV-like rhadinoviruses, RV1 and RV2, has been revealed in Old World primates (including African green monkeys, macaques, and, recently, mandrills), viruses belonging to the RV2 genogroup have not yet been identified from great apes. Indeed, the three yet known ␥2-herpesviruses in chimpanzees (PanRHV1a/PtRV1, PanRHV1b) and gorillas (GorRHV1) belong to the RV1 group. To investigate the putative existence of a new RV2 Rhadinovirus in chimpanzees and gorillas we have used the degenerate consensus primer PCR strategy for the Herpesviral DNA polymerase gene on 40 wild-caught animals. This study led to the discovery, in common chimpanzees, of a novel ␥2-herpesvirus belonging to the RV2 genogroup, termed Pan Rhadino-herpesvirus 2 (PanRHV2). Use of specific primers and internal oligonucleotide probes demonstrated the presence of this novel ␥2-herpesvirus in three wild-caught animals. -

Journal of Virology
JOURNAL OF VIROLOGY Volume 80 March 2006 No. 6 SPOTLIGHT Articles of Significant Interest Selected from This Issue by 2587–2588 the Editors STRUCTURE AND ASSEMBLY Crystal Structure of the Oligomerization Domain of the Haitao Ding, Todd J. Green, 2808–2814 Phosphoprotein of Vesicular Stomatitis Virus Shanyun Lu, and Ming Luo Subcellular Localization of Hepatitis C Virus Structural Yves Rouille´, Franc¸ois Helle, David 2832–2841 Proteins in a Cell Culture System That Efficiently Replicates Delgrange, Philippe Roingeard, the Virus Ce´cile Voisset, Emmanuelle Blanchard, Sandrine Belouzard, Jane McKeating, Arvind H. Patel, Geert Maertens, Takaji Wakita, Czeslaw Wychowski, and Jean Dubuisson A Small Loop in the Capsid Protein of Moloney Murine Marcy R. Auerbach, Kristy R. 2884–2893 Leukemia Virus Controls Assembly of Spherical Cores Brown, Artem Kaplan, Denise de Las Nueces, and Ila R. Singh Identification of the Nucleocapsid, Tegument, and Envelope Jyh-Ming Tsai, Han-Ching Wang, 3021–3029 Proteins of the Shrimp White Spot Syndrome Virus Virion Jiann-Horng Leu, Andrew H.-J. Wang, Ying Zhuang, Peter J. Walker, Guang-Hsiung Kou, and Chu-Fang Lo GENOME REPLICATION AND REGULATION OF VIRAL GENE EXPRESSION Epitope Mapping of Herpes Simplex Virus Type 2 gH/gL Tina M. Cairns, Marie S. Shaner, Yi 2596–2608 Defines Distinct Antigenic Sites, Including Some Associated Zuo, Manuel Ponce-de-Leon, with Biological Function Isabelle Baribaud, Roselyn J. Eisenberg, Gary H. Cohen, and J. Charles Whitbeck The ␣-TIF (VP16) Homologue (ETIF) of Equine Jens von Einem, Daniel 2609–2620 Herpesvirus 1 Is Essential for Secondary Envelopment Schumacher, Dennis J. O’Callaghan, and Virus Egress and Nikolaus Osterrieder Suppression of Viral RNA Recombination by a Host Chi-Ping Cheng, Elena Serviene, 2631–2640 Exoribonuclease and Peter D. -

Comparative Analysis, Distribution, and Characterization of Microsatellites in Orf Virus Genome
www.nature.com/scientificreports OPEN Comparative analysis, distribution, and characterization of microsatellites in Orf virus genome Basanta Pravas Sahu1, Prativa Majee 1, Ravi Raj Singh1, Anjan Sahoo2 & Debasis Nayak 1* Genome-wide in-silico identifcation of microsatellites or simple sequence repeats (SSRs) in the Orf virus (ORFV), the causative agent of contagious ecthyma has been carried out to investigate the type, distribution and its potential role in the genome evolution. We have investigated eleven ORFV strains, which resulted in the presence of 1,036–1,181 microsatellites per strain. The further screening revealed the presence of 83–107 compound SSRs (cSSRs) per genome. Our analysis indicates the dinucleotide (76.9%) repeats to be the most abundant, followed by trinucleotide (17.7%), mononucleotide (4.9%), tetranucleotide (0.4%) and hexanucleotide (0.2%) repeats. The Relative Abundance (RA) and Relative Density (RD) of these SSRs varied between 7.6–8.4 and 53.0–59.5 bp/ kb, respectively. While in the case of cSSRs, the RA and RD ranged from 0.6–0.8 and 12.1–17.0 bp/kb, respectively. Regression analysis of all parameters like the incident of SSRs, RA, and RD signifcantly correlated with the GC content. But in a case of genome size, except incident SSRs, all other parameters were non-signifcantly correlated. Nearly all cSSRs were composed of two microsatellites, which showed no biasedness to a particular motif. Motif duplication pattern, such as, (C)-x-(C), (TG)- x-(TG), (AT)-x-(AT), (TC)- x-(TC) and self-complementary motifs, such as (GC)-x-(CG), (TC)-x-(AG), (GT)-x-(CA) and (TC)-x-(AG) were observed in the cSSRs. -

Characterization of the Rubella Virus Nonstructural Protease Domain and Its Cleavage Site
JOURNAL OF VIROLOGY, July 1996, p. 4707–4713 Vol. 70, No. 7 0022-538X/96/$04.0010 Copyright q 1996, American Society for Microbiology Characterization of the Rubella Virus Nonstructural Protease Domain and Its Cleavage Site 1 2 2 1 JUN-PING CHEN, JAMES H. STRAUSS, ELLEN G. STRAUSS, AND TERYL K. FREY * Department of Biology, Georgia State University, Atlanta, Georgia 30303,1 and Division of Biology, California Institute of Technology, Pasadena, California 911252 Received 27 October 1995/Accepted 3 April 1996 The region of the rubella virus nonstructural open reading frame that contains the papain-like cysteine protease domain and its cleavage site was expressed with a Sindbis virus vector. Cys-1151 has previously been shown to be required for the activity of the protease (L. D. Marr, C.-Y. Wang, and T. K. Frey, Virology 198:586–592, 1994). Here we show that His-1272 is also necessary for protease activity, consistent with the active site of the enzyme being composed of a catalytic dyad consisting of Cys-1151 and His-1272. By means of radiochemical amino acid sequencing, the site in the polyprotein cleaved by the nonstructural protease was found to follow Gly-1300 in the sequence Gly-1299–Gly-1300–Gly-1301. Mutagenesis studies demonstrated that change of Gly-1300 to alanine or valine abrogated cleavage. In contrast, Gly-1299 and Gly-1301 could be changed to alanine with retention of cleavage, but a change to valine abrogated cleavage. Coexpression of a construct that contains a cleavage site mutation (to serve as a protease) together with a construct that contains a protease mutation (to serve as a substrate) failed to reveal trans cleavage. -

Infection Status of Human Parvovirus B19, Cytomegalovirus and Herpes Simplex Virus-1/2 in Women with First-Trimester Spontaneous
Gao et al. Virology Journal (2018) 15:74 https://doi.org/10.1186/s12985-018-0988-5 RESEARCH Open Access Infection status of human parvovirus B19, cytomegalovirus and herpes simplex Virus- 1/2 in women with first-trimester spontaneous abortions in Chongqing, China Ya-Ling Gao1, Zhan Gao3,4, Miao He3,4* and Pu Liao2* Abstract Background: Infection with Parvovirus B19 (B19V), Cytomegalovirus (CMV) and Herpes Simplex Virus-1/2 (HSV-1/2) may cause fetal loses including spontaneous abortion, intrauterine fetal death and non-immune hydrops fetalis. Few comprehensive studies have investigated first-trimester spontaneous abortions caused by virus infections in Chongqing, China. Our study intends to investigate the infection of B19V, CMV and HSV-1/2 in first-trimester spontaneous abortions and the corresponding immune response. Methods: 100 abortion patients aged from 17 to 47 years were included in our study. The plasma samples (100) were analyzed qualitatively for specific IgG/IgM for B19V, CMV and HSV-1/2 (Virion\Serion, Germany) according to the manufacturer’s recommendations. B19V, CMV and HSV-1/2 DNA were quantification by Real-Time PCR. Results: No specimens were positive for B19V, CMV, and HSV-1/2 DNA. By serology, 30.0%, 95.0%, 92.0% of patients were positive for B19V, CMV and HSV-1/2 IgG respectively, while 2% and 1% for B19V and HSV-1/2 IgM. Conclusion: The low rate of virus DNA and a high proportion of CMV and HSV-1/2 IgG for most major of abortion patients in this study suggest that B19V, CMV and HSV-1/2 may not be the common factor leading to the spontaneous abortion of early pregnancy. -

Measles and Rubella Initiative Outbreak Response Fund Application Standard Operating Procedures
M&RI SOP Feb 24, 2020 Final Measles and Rubella Initiative Outbreak Response Fund Application Standard Operating Procedures This update of the M&RI ORF Standard Operating Procedures (SOPs) includes the following modifications: 1. To encourage countries to submit applications for ORF support early in the evolution of the outbreak in order to effectively stop measles virus transmission before it becomes more widespread, M&RI will monitor indicators of timeliness of outbreak response immunization in accordance with Immunization Agenda 2030 guidelines; 2. To accelerate the process to receive support, M&RI has removed requirements for advance notification when requesting ORF support and has established a limited timeframe to evaluate the application for ORF support and to provide feedback or issue a decision letter; 3. To more comprehensively address outbreak response linked to timely and efficient use of vaccine, M&RI will allow for greater flexibility in areas of support on an exceptional basis and with adequate justification; 4. To reduce the likelihood of requested additional information or clarifications from M&RI, the SOPs provide greater detail regarding the key data elements and analysis recommended when investigating measles outbreaks and preparing reports, plans and budgets; 5. To maximize use of outbreak investigations and their follow up to strengthen immunization systems, M&RI requests countries to include findings from root cause analyses and, based on these, plans to improve routine immunization, surveillance and outbreak preparedness in the required post-outbreak report. A. Background The Measles and Rubella Initiative (M&RI) has provided funding through an outbreak response fund (ORF) since 2012 to support bundled vaccine and operational costs for measles and rubella outbreak response immunization (ORI), with Gavi supporting up to a total of US$10 million per year for Gavi-eligible countries. -

Chlamydia Trachomatis Infection Is Driven by Nonprotective Immune Cells That Are Distinct from Protective Populations
Pathology after Chlamydia trachomatis infection is driven by nonprotective immune cells that are distinct from protective populations Rebeccah S. Lijeka,b,1, Jennifer D. Helblea, Andrew J. Olivea,c, Kyra W. Seigerb, and Michael N. Starnbacha,1 aDepartment of Microbiology and Immunobiology, Harvard Medical School, Boston, MA 02115; bDepartment of Biological Sciences, Mount Holyoke College, South Hadley, MA 01075; and cDepartment of Microbiology and Physiological Systems, University of Massachusetts Medical School, Worcester, MA 01605 Edited by Rafi Ahmed, Emory University, Atlanta, GA, and approved December 27, 2017 (received for review June 23, 2017) Infection with Chlamydia trachomatis drives severe mucosal immu- sequence identity, Chlamydia muridarum, the extent to which the nopathology; however, the immune responses that are required for molecular pathogenesis of C. muridarum represents that of Ct is mediating pathology vs. protection are not well understood. Here, unknown (6). Ct serovar L2 (Ct L2) is capable of infecting the we employed a mouse model to identify immune responses re- mouse upper genital tract when inoculated across the cervix into quired for C. trachomatis-induced upper genital tract pathology the uterus (7, 8) but it does not induce robust immunopathology. and to determine whether these responses are also required for This is consistent with the human disease phenotype caused by Ct L2, bacterial clearance. In mice as in humans, immunopathology was which disseminates to the lymph nodes causing lymphogranuloma characterized by extravasation of leukocytes into the upper genital venereum (LGV) and is not a major cause of mucosal immunopa- thology in the female upper genital tract (uterus and ovaries). tract that occluded luminal spaces in the uterus and ovaries. -

Where Do We Stand After Decades of Studying Human Cytomegalovirus?
microorganisms Review Where do we Stand after Decades of Studying Human Cytomegalovirus? 1, 2, 1 1 Francesca Gugliesi y, Alessandra Coscia y, Gloria Griffante , Ganna Galitska , Selina Pasquero 1, Camilla Albano 1 and Matteo Biolatti 1,* 1 Laboratory of Pathogenesis of Viral Infections, Department of Public Health and Pediatric Sciences, University of Turin, 10126 Turin, Italy; [email protected] (F.G.); gloria.griff[email protected] (G.G.); [email protected] (G.G.); [email protected] (S.P.); [email protected] (C.A.) 2 Complex Structure Neonatology Unit, Department of Public Health and Pediatric Sciences, University of Turin, 10126 Turin, Italy; [email protected] * Correspondence: [email protected] These authors contributed equally to this work. y Received: 19 March 2020; Accepted: 5 May 2020; Published: 8 May 2020 Abstract: Human cytomegalovirus (HCMV), a linear double-stranded DNA betaherpesvirus belonging to the family of Herpesviridae, is characterized by widespread seroprevalence, ranging between 56% and 94%, strictly dependent on the socioeconomic background of the country being considered. Typically, HCMV causes asymptomatic infection in the immunocompetent population, while in immunocompromised individuals or when transmitted vertically from the mother to the fetus it leads to systemic disease with severe complications and high mortality rate. Following primary infection, HCMV establishes a state of latency primarily in myeloid cells, from which it can be reactivated by various inflammatory stimuli. Several studies have shown that HCMV, despite being a DNA virus, is highly prone to genetic variability that strongly influences its replication and dissemination rates as well as cellular tropism. In this scenario, the few currently available drugs for the treatment of HCMV infections are characterized by high toxicity, poor oral bioavailability, and emerging resistance. -

Characterization of Host Micrornas That Respond to DNA Virus Infection in a Crustacean Tianzhi Huang, Dandan Xu and Xiaobo Zhang*
Huang et al. BMC Genomics 2012, 13:159 http://www.biomedcentral.com/1471-2164/13/159 RESEARCH ARTICLE Open Access Characterization of host microRNAs that respond to DNA virus infection in a crustacean Tianzhi Huang, Dandan Xu and Xiaobo Zhang* Abstract Background: MicroRNAs (miRNAs) are key posttranscriptional regulators of gene expression that are implicated in many processes of eukaryotic cells. It is known that the expression profiles of host miRNAs can be reshaped by viruses. However, a systematic investigation of marine invertebrate miRNAs that respond to virus infection has not yet been performed. Results: In this study, the shrimp Marsupenaeus japonicus was challenged by white spot syndrome virus (WSSV). Small RNA sequencing of WSSV-infected shrimp at different time post-infection (0, 6, 24 and 48 h) identified 63 host miRNAs, 48 of which were conserved in other animals, representing 43 distinct families. Of the identified host miRNAs, 31 were differentially expressed in response to virus infection, of which 25 were up-regulated and six down-regulated. The results were confirmed by northern blots. The TargetScan and miRanda algorithms showed that most target genes of the differentially expressed miRNAs were related to immune responses. Gene ontology analysis revealed that immune signaling pathways were mediated by these miRNAs. Evolutionary analysis showed that three of them, miR-1, miR-7 and miR-34, are highly conserved in shrimp, fruit fly and humans and function in the similar pathways. Conclusions: Our study provides the first large-scale characterization of marine invertebrate miRNAs that respond to virus infection. This will help to reveal the molecular events involved in virus-host interactions mediated by miRNAs and their evolution in animals. -

Leptospirosis: a Waterborne Zoonotic Disease of Global Importance
August 2006 volume 22 number 08 Leptospirosis: A waterborne zoonotic disease of global importance INTRODUCTION syndrome has two phases: a septicemic and an immune phase (Levett, 2005). Leptospirosis is considered one of the most common zoonotic diseases It is in the immune phase that organ-specific damage and more severe illness globally. In the United States, outbreaks are increasingly being reported is seen. See text box for more information on the two phases. The typical among those participating in recreational water activities (Centers for Disease presenting signs of leptospirosis in humans are fever, headache, chills, con- Control and Prevention [CDC], 1996, 1998, and 2001) and sporadic cases are junctival suffusion, and myalgia (particularly in calf and lumbar areas) often underdiagnosed. With the onset of warm temperatures, increased (Heymann, 2004). Less common signs include a biphasic fever, meningitis, outdoor activities, and travel, Georgia may expect to see more leptospirosis photosensitivity, rash, and hepatic or renal failure. cases. DIAGNOSIS OF LEPTOSPIROSIS Leptospirosis is a zoonosis caused by infection with the bacterium Leptospira Detecting serum antibodies against leptospira interrogans. The disease occurs worldwide, but it is most common in temper- • Microscopic Agglutination Titers (MAT) ate regions in the late summer and early fall and in tropical regions during o Paired serum samples which show a four-fold rise in rainy seasons. It is not surprising that Hawaii has the highest incidence of titer confirm the diagnosis; a single high titer in a per- leptospirosis in the United States (Levett, 2005). The reservoir of pathogenic son clinically suspected to have leptospirosis is highly leptospires is the renal tubules of wild and domestic animals. -

Gonorrhea, Chlamydia, and Syphilis
2019 GONORRHEA, CHLAMYDIA, AND SYPHILIS AND CHLAMYDIA, GONORRHEA, Dedication TAG would like to thank the National Coalition of STD Directors for funding and input on the report. THE PIPELINE REPORT Pipeline for Gonorrhea, Chlamydia, and Syphilis By Jeremiah Johnson Introduction The current toolbox for addressing gonorrhea, chlamydia, and syphilis is inadequate. At a time where all three epidemics are dramatically expanding in locations all around the globe, including record-breaking rates of new infections in the United States, stakeholders must make do with old tools, inadequate systems for addressing sexual health, and a sparse research pipeline of new treatment, prevention, and diagnostic options. Lack of investment in sexual health research has left the field with inadequate prevention options, and limited access to infrastructure for testing and treatment have allowed sexually transmitted infections (STIs) to flourish. The consequences of this underinvestment are large: according to the World Health Organization (WHO), in 2012 there were an estimated 357 million new infections (roughly 1 million per day) of the four curable STIs: gonorrhea, chlamydia, syphilis, and trichomoniasis.1 In the United States, the three reportable STIs that are the focus of this report—gonorrhea, chlamydia, and syphilis—are growing at record paces. In 2017, a total of 30,644 cases of primary and secondary (P&S) syphilis—the most infectious stages of the disease—were reported in the United States. Since reaching a historic low in 2000 and 2001, the rate of P&S syphilis has increased almost every year, increasing 10.5% during 2016–2017. Also in 2017, 555,608 cases of gonorrhea were reported to the U.S. -
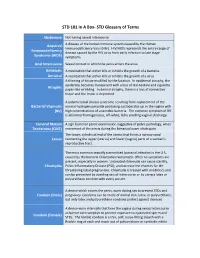
STD Glossary of Terms
STD 101 In A Box- STD Glossary of Terms Abstinence Not having sexual intercourse Acquired A disease of the human immune system caused by the Human Immunodeficiency Virus (HIV). HIV/AIDS represents the entire range of Immunodeficiency disease caused by the HIV virus from early infection to late stage Syndrome (AIDS) symptoms. Anal Intercourse Sexual contact in which the penis enters the anus. Antibiotic A medication that either kills or inhibits the growth of a bacteria. Antiviral A medication that either kills or inhibits the growth of a virus. A thinning of tissue modified by the location. In epidermal atrophy, the epidermis becomes transparent with a loss of skin texture and cigarette Atrophic paper-like wrinkling. In dermal atrophy, there is a loss of connective tissue and the lesion is depressed. A polymicrobial clinical syndrome resulting from replacement of the Bacterial Vaginosis normal hydrogen peroxide producing Lactobacillus sp. in the vagina with (BV) high concentrations of anaerobic bacteria. The common symptom of BV is abnormal homogeneous, off-white, fishy smelling vaginal discharge. Cervical Motion A sign found on pelvic examination suggestive of pelvic pathology; when Tenderness (CMT) movement of the cervix during the bimanual exam elicits pain. The lower, cylindrical end of the uterus that forms a narrow canal Cervix connecting the upper (uterus) and lower (vagina) parts of a woman's reproductive tract. The most common sexually transmitted bacterial infection in the U.S., caused by the bacteria Chlamydia trachomatis. Often no symptoms are present, especially in women. Untreated chlamydia can cause sterility, Chlamydia Pelvic Inflammatory Disease (PID), and increase the chances for life- threatening tubal pregnancies.