Chlamydia Trachomatis and Chlamydia Pneumoniae Interaction with the Host: Latest Advances and Future Prospective
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Compendium of Measures to Control Chlamydia Psittaci Infection Among
Compendium of Measures to Control Chlamydia psittaci Infection Among Humans (Psittacosis) and Pet Birds (Avian Chlamydiosis), 2017 Author(s): Gary Balsamo, DVM, MPH&TMCo-chair Angela M. Maxted, DVM, MS, PhD, Dipl ACVPM Joanne W. Midla, VMD, MPH, Dipl ACVPM Julia M. Murphy, DVM, MS, Dipl ACVPMCo-chair Ron Wohrle, DVM Thomas M. Edling, DVM, MSpVM, MPH (Pet Industry Joint Advisory Council) Pilar H. Fish, DVM (American Association of Zoo Veterinarians) Keven Flammer, DVM, Dipl ABVP (Avian) (Association of Avian Veterinarians) Denise Hyde, PharmD, RP Preeta K. Kutty, MD, MPH Miwako Kobayashi, MD, MPH Bettina Helm, DVM, MPH Brit Oiulfstad, DVM, MPH (Council of State and Territorial Epidemiologists) Branson W. Ritchie, DVM, MS, PhD, Dipl ABVP, Dipl ECZM (Avian) Mary Grace Stobierski, DVM, MPH, Dipl ACVPM (American Veterinary Medical Association Council on Public Health and Regulatory Veterinary Medicine) Karen Ehnert, and DVM, MPVM, Dipl ACVPM (American Veterinary Medical Association Council on Public Health and Regulatory Veterinary Medicine) Thomas N. Tully JrDVM, MS, Dipl ABVP (Avian), Dipl ECZM (Avian) (Association of Avian Veterinarians) Source: Journal of Avian Medicine and Surgery, 31(3):262-282. Published By: Association of Avian Veterinarians https://doi.org/10.1647/217-265 URL: http://www.bioone.org/doi/full/10.1647/217-265 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. -

What Is Lyme Disease?
The threat of Lyme disease Dr Roger Evans Consultant Clinical Scientist National Lyme Borreliosis Testing Laboratory What is Lyme disease? • Also known as Lyme borreliosis • An infection caused by Borrelia burgdorferi sensu lato, a gram negative spirochaete bacterium • Transmitted through the bite of an infected tick (Ixodes ricinus in the UK) – a tick needs to be attached to the skin for around 24 hours to transmit Borrelia sp. to a person. • Recognised clinical presentations: early localized, early disseminated and late Lyme disease • There are large gaps in our clinical understanding of Lyme disease, particularly for ‘Post Treatment Lyme Disease Syndrome ‘ Picture credit: James Gathany Ixodes ricinus (tick) ecology/ life cycle Adapted from Manelli et al. 2011 Laboratory samples and cases of LB in Scotland 1996 to 2016 500 7000 450 6000 400 350 5000 300 4000 250 3000 200 Number of cases Number of samples 150 2000 100 1000 50 0 0 Cases Seroneg EM Year Samples GP study (2010-2012) • Three GP practices in NHS Highland region: Nairn, Culloden, Fort William • Number of laboratory cases compared to number of cases treated for Lyme disease without testing • About 2 x number of cases diagnosed by GP without lab testing compared to those that were tested • 2010-2011 • 1440 blood donors • Screened by EIA • EIA positive or equivocal samples confirmed by immunoblot (IB) • 60/1440 (4.2%) IB positive Munro et al (2015) Transfusion Medicine. Letter to the Editor doi: 10.1111/tme.12197 What does it cost? • Netherlands study (2010) – 5 health outcomes: -

Chlamydia Trachomatis Infection Is Driven by Nonprotective Immune Cells That Are Distinct from Protective Populations
Pathology after Chlamydia trachomatis infection is driven by nonprotective immune cells that are distinct from protective populations Rebeccah S. Lijeka,b,1, Jennifer D. Helblea, Andrew J. Olivea,c, Kyra W. Seigerb, and Michael N. Starnbacha,1 aDepartment of Microbiology and Immunobiology, Harvard Medical School, Boston, MA 02115; bDepartment of Biological Sciences, Mount Holyoke College, South Hadley, MA 01075; and cDepartment of Microbiology and Physiological Systems, University of Massachusetts Medical School, Worcester, MA 01605 Edited by Rafi Ahmed, Emory University, Atlanta, GA, and approved December 27, 2017 (received for review June 23, 2017) Infection with Chlamydia trachomatis drives severe mucosal immu- sequence identity, Chlamydia muridarum, the extent to which the nopathology; however, the immune responses that are required for molecular pathogenesis of C. muridarum represents that of Ct is mediating pathology vs. protection are not well understood. Here, unknown (6). Ct serovar L2 (Ct L2) is capable of infecting the we employed a mouse model to identify immune responses re- mouse upper genital tract when inoculated across the cervix into quired for C. trachomatis-induced upper genital tract pathology the uterus (7, 8) but it does not induce robust immunopathology. and to determine whether these responses are also required for This is consistent with the human disease phenotype caused by Ct L2, bacterial clearance. In mice as in humans, immunopathology was which disseminates to the lymph nodes causing lymphogranuloma characterized by extravasation of leukocytes into the upper genital venereum (LGV) and is not a major cause of mucosal immunopa- thology in the female upper genital tract (uterus and ovaries). tract that occluded luminal spaces in the uterus and ovaries. -

Table S4. Phylogenetic Distribution of Bacterial and Archaea Genomes in Groups A, B, C, D, and X
Table S4. Phylogenetic distribution of bacterial and archaea genomes in groups A, B, C, D, and X. Group A a: Total number of genomes in the taxon b: Number of group A genomes in the taxon c: Percentage of group A genomes in the taxon a b c cellular organisms 5007 2974 59.4 |__ Bacteria 4769 2935 61.5 | |__ Proteobacteria 1854 1570 84.7 | | |__ Gammaproteobacteria 711 631 88.7 | | | |__ Enterobacterales 112 97 86.6 | | | | |__ Enterobacteriaceae 41 32 78.0 | | | | | |__ unclassified Enterobacteriaceae 13 7 53.8 | | | | |__ Erwiniaceae 30 28 93.3 | | | | | |__ Erwinia 10 10 100.0 | | | | | |__ Buchnera 8 8 100.0 | | | | | | |__ Buchnera aphidicola 8 8 100.0 | | | | | |__ Pantoea 8 8 100.0 | | | | |__ Yersiniaceae 14 14 100.0 | | | | | |__ Serratia 8 8 100.0 | | | | |__ Morganellaceae 13 10 76.9 | | | | |__ Pectobacteriaceae 8 8 100.0 | | | |__ Alteromonadales 94 94 100.0 | | | | |__ Alteromonadaceae 34 34 100.0 | | | | | |__ Marinobacter 12 12 100.0 | | | | |__ Shewanellaceae 17 17 100.0 | | | | | |__ Shewanella 17 17 100.0 | | | | |__ Pseudoalteromonadaceae 16 16 100.0 | | | | | |__ Pseudoalteromonas 15 15 100.0 | | | | |__ Idiomarinaceae 9 9 100.0 | | | | | |__ Idiomarina 9 9 100.0 | | | | |__ Colwelliaceae 6 6 100.0 | | | |__ Pseudomonadales 81 81 100.0 | | | | |__ Moraxellaceae 41 41 100.0 | | | | | |__ Acinetobacter 25 25 100.0 | | | | | |__ Psychrobacter 8 8 100.0 | | | | | |__ Moraxella 6 6 100.0 | | | | |__ Pseudomonadaceae 40 40 100.0 | | | | | |__ Pseudomonas 38 38 100.0 | | | |__ Oceanospirillales 73 72 98.6 | | | | |__ Oceanospirillaceae -

Leptospirosis: a Waterborne Zoonotic Disease of Global Importance
August 2006 volume 22 number 08 Leptospirosis: A waterborne zoonotic disease of global importance INTRODUCTION syndrome has two phases: a septicemic and an immune phase (Levett, 2005). Leptospirosis is considered one of the most common zoonotic diseases It is in the immune phase that organ-specific damage and more severe illness globally. In the United States, outbreaks are increasingly being reported is seen. See text box for more information on the two phases. The typical among those participating in recreational water activities (Centers for Disease presenting signs of leptospirosis in humans are fever, headache, chills, con- Control and Prevention [CDC], 1996, 1998, and 2001) and sporadic cases are junctival suffusion, and myalgia (particularly in calf and lumbar areas) often underdiagnosed. With the onset of warm temperatures, increased (Heymann, 2004). Less common signs include a biphasic fever, meningitis, outdoor activities, and travel, Georgia may expect to see more leptospirosis photosensitivity, rash, and hepatic or renal failure. cases. DIAGNOSIS OF LEPTOSPIROSIS Leptospirosis is a zoonosis caused by infection with the bacterium Leptospira Detecting serum antibodies against leptospira interrogans. The disease occurs worldwide, but it is most common in temper- • Microscopic Agglutination Titers (MAT) ate regions in the late summer and early fall and in tropical regions during o Paired serum samples which show a four-fold rise in rainy seasons. It is not surprising that Hawaii has the highest incidence of titer confirm the diagnosis; a single high titer in a per- leptospirosis in the United States (Levett, 2005). The reservoir of pathogenic son clinically suspected to have leptospirosis is highly leptospires is the renal tubules of wild and domestic animals. -

Gonorrhea, Chlamydia, and Syphilis
2019 GONORRHEA, CHLAMYDIA, AND SYPHILIS AND CHLAMYDIA, GONORRHEA, Dedication TAG would like to thank the National Coalition of STD Directors for funding and input on the report. THE PIPELINE REPORT Pipeline for Gonorrhea, Chlamydia, and Syphilis By Jeremiah Johnson Introduction The current toolbox for addressing gonorrhea, chlamydia, and syphilis is inadequate. At a time where all three epidemics are dramatically expanding in locations all around the globe, including record-breaking rates of new infections in the United States, stakeholders must make do with old tools, inadequate systems for addressing sexual health, and a sparse research pipeline of new treatment, prevention, and diagnostic options. Lack of investment in sexual health research has left the field with inadequate prevention options, and limited access to infrastructure for testing and treatment have allowed sexually transmitted infections (STIs) to flourish. The consequences of this underinvestment are large: according to the World Health Organization (WHO), in 2012 there were an estimated 357 million new infections (roughly 1 million per day) of the four curable STIs: gonorrhea, chlamydia, syphilis, and trichomoniasis.1 In the United States, the three reportable STIs that are the focus of this report—gonorrhea, chlamydia, and syphilis—are growing at record paces. In 2017, a total of 30,644 cases of primary and secondary (P&S) syphilis—the most infectious stages of the disease—were reported in the United States. Since reaching a historic low in 2000 and 2001, the rate of P&S syphilis has increased almost every year, increasing 10.5% during 2016–2017. Also in 2017, 555,608 cases of gonorrhea were reported to the U.S. -
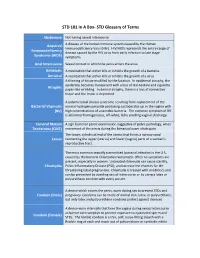
STD Glossary of Terms
STD 101 In A Box- STD Glossary of Terms Abstinence Not having sexual intercourse Acquired A disease of the human immune system caused by the Human Immunodeficiency Virus (HIV). HIV/AIDS represents the entire range of Immunodeficiency disease caused by the HIV virus from early infection to late stage Syndrome (AIDS) symptoms. Anal Intercourse Sexual contact in which the penis enters the anus. Antibiotic A medication that either kills or inhibits the growth of a bacteria. Antiviral A medication that either kills or inhibits the growth of a virus. A thinning of tissue modified by the location. In epidermal atrophy, the epidermis becomes transparent with a loss of skin texture and cigarette Atrophic paper-like wrinkling. In dermal atrophy, there is a loss of connective tissue and the lesion is depressed. A polymicrobial clinical syndrome resulting from replacement of the Bacterial Vaginosis normal hydrogen peroxide producing Lactobacillus sp. in the vagina with (BV) high concentrations of anaerobic bacteria. The common symptom of BV is abnormal homogeneous, off-white, fishy smelling vaginal discharge. Cervical Motion A sign found on pelvic examination suggestive of pelvic pathology; when Tenderness (CMT) movement of the cervix during the bimanual exam elicits pain. The lower, cylindrical end of the uterus that forms a narrow canal Cervix connecting the upper (uterus) and lower (vagina) parts of a woman's reproductive tract. The most common sexually transmitted bacterial infection in the U.S., caused by the bacteria Chlamydia trachomatis. Often no symptoms are present, especially in women. Untreated chlamydia can cause sterility, Chlamydia Pelvic Inflammatory Disease (PID), and increase the chances for life- threatening tubal pregnancies. -

Chlamydial Genital Infection(Chlamydia Trachomatis)
Chlamydial Genital Infection (Chlamydia trachomatis) February 2003 1) THE DISEASE AND ITS EPIDEMIOLOGY A. Etiologic Agent Chlamydial genital infection (CGI) is caused by the obligate, intracellular bacterium Chlamydia trachomatis immunotypes D through K. B. Clinical Description and Laboratory Diagnosis A sexually transmitted genital infection that manifests in males primarily as urethritis and in females as mucopurulent cervicitis. Clinical manifestations are difficult to distinguish from gonorrhea. Males may present with a mucopurulent discharges of scanty to moderate quantity, urethral itching and dysuria. Asymptomatic infection may be found in 1%-25% of sexually active men. Possible complications include epididymitis, infertility and Reiter syndrome. Anorectal intercourse may result in chlamydial proctitis. Women frequently present with a mucopurulent endocervical discharge including edema, erythema and easily induced endocervical bleeding. However, most women with endocervical or urethral infections are asymptomatic. Possible complications include salpingitis with subsequent risk of infertility and ectopic pregnancy. Asymptomatic chronic infections of the endometrium and fallopian tubes may lead to the same outcomes. Less frequent manifestations include bartholinitis, urethral syndrome with dysuria and pyuria, perihepatitis (Fitz-Hugh-Curtis syndrome), and proctitis. Infection during pregnancy may result in premature rupture of membranes and preterm delivery and conjunctival and pneumonic infection of the newborn. Laboratory diagnosis is based upon the identification of Chlamydia in intraurethral or endocervical smear by direct immunofluorescence test, enzyme immunoassay, DNA probe, and nucleic acid amplification test (NAAT) or cell culture. NAAT can be used with urine specimens. C. Vectors and Reservoirs Humans. D. Modes of Transmission By sexual contact and through perinatal exposure to the mother’s infected cervix. -

Chlamydia, Gonorrhoea, Trichomoniasis and Syphilis
Research Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016 Jane Rowley,a Stephen Vander Hoorn,b Eline Korenromp,c Nicola Low,d Magnus Unemo,e Laith J Abu- Raddad,f R Matthew Chico,g Alex Smolak,f Lori Newman,h Sami Gottlieb,a Soe Soe Thwin,a Nathalie Brouteta & Melanie M Taylora Objective To generate estimates of the global prevalence and incidence of urogenital infection with chlamydia, gonorrhoea, trichomoniasis and syphilis in women and men, aged 15–49 years, in 2016. Methods For chlamydia, gonorrhoea and trichomoniasis, we systematically searched for studies conducted between 2009 and 2016 reporting prevalence. We also consulted regional experts. To generate estimates, we used Bayesian meta-analysis. For syphilis, we aggregated the national estimates generated by using Spectrum-STI. Findings For chlamydia, gonorrhoea and/or trichomoniasis, 130 studies were eligible. For syphilis, the Spectrum-STI database contained 978 data points for the same period. The 2016 global prevalence estimates in women were: chlamydia 3.8% (95% uncertainty interval, UI: 3.3–4.5); gonorrhoea 0.9% (95% UI: 0.7–1.1); trichomoniasis 5.3% (95% UI:4.0–7.2); and syphilis 0.5% (95% UI: 0.4–0.6). In men prevalence estimates were: chlamydia 2.7% (95% UI: 1.9–3.7); gonorrhoea 0.7% (95% UI: 0.5–1.1); trichomoniasis 0.6% (95% UI: 0.4–0.9); and syphilis 0.5% (95% UI: 0.4–0.6). Total estimated incident cases were 376.4 million: 127.2 million (95% UI: 95.1–165.9 million) chlamydia cases; 86.9 million (95% UI: 58.6–123.4 million) gonorrhoea cases; 156.0 million (95% UI: 103.4–231.2 million) trichomoniasis cases; and 6.3 million (95% UI: 5.5–7.1 million) syphilis cases. -

Genetic Diversity in Treponema Pallidum: Implications for Pathogenesis, Evolution and Molecular Diagnostics of Syphilis and Yaws ⇑ David Šmajs A, , Steven J
Infection, Genetics and Evolution 12 (2012) 191–202 Contents lists available at SciVerse ScienceDirect Infection, Genetics and Evolution journal homepage: www.elsevier.com/locate/meegid Review Genetic diversity in Treponema pallidum: Implications for pathogenesis, evolution and molecular diagnostics of syphilis and yaws ⇑ David Šmajs a, , Steven J. Norris b, George M. Weinstock c a Department of Biology, Faculty of Medicine, Masaryk University, Kamenice 5, Building A6, 625 00 Brno, Czech Republic b Department of Pathology and Laboratory Medicine, University of Texas Medical School at Houston, 6431 Fannin Street, Houston, TX 77030, USA c The Genome Institute, Washington University, 4444 Forest Park Avenue, Campus Box 8501, St. Louis, MO 63108, USA article info abstract Article history: Pathogenic uncultivable treponemes, similar to syphilis-causing Treponema pallidum subspecies pallidum, Received 21 September 2011 include T. pallidum ssp. pertenue, T. pallidum ssp. endemicum and Treponema carateum, which cause yaws, Received in revised form 5 December 2011 bejel and pinta, respectively. Genetic analyses of these pathogens revealed striking similarity among Accepted 7 December 2011 these bacteria and also a high degree of similarity to the rabbit pathogen, Treponema paraluiscuniculi,a Available online 15 December 2011 treponeme not infectious to humans. Genome comparisons between pallidum and non-pallidum trepo- nemes revealed genes with potential involvement in human infectivity, whereas comparisons between Keywords: pallidum and pertenue treponemes identified genes possibly involved in the high invasivity of syphilis Treponema pallidum treponemes. Genetic variability within syphilis strains is considered as the basis of syphilis molecular Treponema pallidum ssp. pertenue Treponema pallidum ssp. endemicum epidemiology with potential to detect more virulent strains, whereas genetic variability within a single Treponema paraluiscuniculi strain is related to its ability to elude the immune system of the host. -

Chlamydia Pneumoniae Is Present in the Dental Plaque of Periodontitis
University of the Pacific Scholarly Commons Dugoni School of Dentistry Faculty Articles Arthur A. Dugoni School of Dentistry 3-11-2019 Chlamydia pneumoniae is present in the dental plaque of periodontitis patients and stimulates an inflammatory response in gingival epithelial cells Cássio Luiz Coutinho Almeida-da-Silva Tamer Alpagot Ye Zhu Sonho Sierra Lee Brian P Roberts See next page for additional authors Follow this and additional works at: https://scholarlycommons.pacific.edu/dugoni-facarticles Part of the Dentistry Commons Recommended Citation Almeida-da-Silva, C., Alpagot, T., Zhu, Y., Lee, S., Roberts, B., Hung, S., Tang, N., & Ojcius, D. (2019). Chlamydia pneumoniae is present in the dental plaque of periodontitis patients and stimulates an inflammatory response in gingival epithelial cells. Microbial Cell, 6(4), 197–208. DOI: 10.15698/mic2019.04.674 https://scholarlycommons.pacific.edu/dugoni-facarticles/459 This Article is brought to you for free and open access by the Arthur A. Dugoni School of Dentistry at Scholarly Commons. It has been accepted for inclusion in Dugoni School of Dentistry Faculty Articles by an authorized administrator of Scholarly Commons. For more information, please contact [email protected]. Authors Cássio Luiz Coutinho Almeida-da-Silva, Tamer Alpagot, Ye Zhu, Sonho Sierra Lee, Brian P Roberts, Shu- Chen Hung, Norina Tang, and David M Ojcius This article is available at Scholarly Commons: https://scholarlycommons.pacific.edu/dugoni-facarticles/459 Research Article www.microbialcell.com Chlamydia pneumoniae is present in the dental plaque of periodontitis patients and stimulates an inflammatory response in gingival epithelial cells Cássio Luiz Coutinho Almeida-da-Silva1, Tamer Alpagot2, Ye Zhu1, Sonho Sierra Lee3,4, Brian P. -

Evaluation and Determination of the Sensitivity and Specificity of a Treponema Pallidum Dried Blood Spot Method for Serologic Diagnosis of Syphilis
Georgia State University ScholarWorks @ Georgia State University Public Health Theses School of Public Health Fall 12-20-2012 Evaluation and Determination of the Sensitivity and Specificity of a Treponema Pallidum Dried Blood Spot Method for Serologic Diagnosis of Syphilis David K. Turgeon Follow this and additional works at: https://scholarworks.gsu.edu/iph_theses Recommended Citation Turgeon, David K., "Evaluation and Determination of the Sensitivity and Specificity of a rT eponema Pallidum Dried Blood Spot Method for Serologic Diagnosis of Syphilis." Thesis, Georgia State University, 2012. https://scholarworks.gsu.edu/iph_theses/239 This Thesis is brought to you for free and open access by the School of Public Health at ScholarWorks @ Georgia State University. It has been accepted for inclusion in Public Health Theses by an authorized administrator of ScholarWorks @ Georgia State University. For more information, please contact [email protected]. Institute of Public Health Public Health Thesis Georgia State University Year 2012 EVALUATION AND DETERMINATION OF THE SENSITIVITY AND SPECIFICITY OF A Treponema pallidum DRIED BLOOD SPOT METHOD FOR SEROLOGIC DIAGNOSIS OF SYPHILIS David K. Turgeon Georgia State University, [email protected] ii ABSTRACT EVALUATION AND DETERMINATION OF THE SENSITIVITY AND SPECIFICITY OF A Treponema pallidum DRIED BLOOD SPOT (DBS) METHOD FOR SEROLOGIC DIAGNOSIS OF SYPHILIS Background: Syphilis is a sexually transmitted infection (STI) caused by Treponema pallidum subspecies pallidum. Syphilis is known as the “great imitator" due to the similarity of clinical signs and symptoms to other infectious diseases. The primary diagnosis of syphilis relies on clinical findings, including the examination of treponemal lesions, and/or serologic tests.