2008 LASKER AWARDS for MEDICAL RESEARCH
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence from Covid-19 Pandemic
RISK PERCEPTIONS AND PROTECTIVE BEHAVIORS: EVIDENCE FROM COVID-19 PANDEMIC Maria Polyakova M. Kate Bundorf Duke University Stanford University & NBER & NBER Jill DeMatteis Jialu L. Streeter Westat Stanford University Grant Miller Jonathan Wivagg Stanford University Westat & NBER April, 2021 Working Paper No. 21-022 Risk Perceptions and Protective Behaviors: Evidence from COVID-19 Pandemic.∗ M. Kate Bundorf Jill DeMatteis Grant Miller Maria Polyakova Duke University Westat Stanford University Stanford University and NBER and NBER and NBER Jialu L. Streeter Jonathan Wivagg Stanford University Westat April 28, 2021 Abstract We analyze data from a survey we administered during the COVID-19 pandemic to investigate the relationship between people's subjective risk beliefs and their protective behaviors. We report three main findings. First, on average, people substantially overestimate the absolute level of risk associated with economic activity, but have correct signals about their relative risk. Second, people who believe that they face a higher risk of infection are more likely to report avoiding economic activities. Third, government mandates restricting economic behavior attenuate the relationship between subjective risk beliefs and protective behaviors. ∗Corresponding author: Maria Polyakova ([email protected]). We are grateful to Sarah B¨ogland Aava Farhadi for excellent research assistance. We have made the de-identified survey dataset used in this manuscript available at the following link. We gratefully acknowledge in-kind survey support from Westat. 1 1 Introduction Subjective perceptions of risk guide decision making in nearly all economic domains (Hurd, 2009). While a rich theoretical literature highlights the importance of subjective beliefs for individual ac- tions, fewer studies provide empirical evidence of the link between risk perceptions and behaviors, particularly in non-laboratory settings and in high-income countries (Peltzman, 1975; Delavande, 2014; Delavande and Kohler, 2015; Mueller et al., 2021). -
19 International Workshop on Low Temperature Detectors
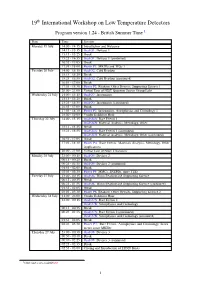
19th International Workshop on Low Temperature Detectors Program version 1.24 - British Summer Time 1 Date Time Session Monday 19 July 14:00 - 14:15 Introduction and Welcome 14:15 - 15:15 Oral O1: Devices 1 15:15 - 15:25 Break 15:25 - 16:55 Oral O1: Devices 1 (continued) 16:55 - 17:05 Break 17:05 - 18:00 Poster P1: MKIDs and TESs 1 Tuesday 20 July 14:00 - 15:15 Oral O2: Cold Readout 15:15 - 15:25 Break 15:25 - 16:55 Oral O2: Cold Readout (continued) 16:55 - 17:05 Break 17:05 - 18:30 Poster P2: Readout, Other Devices, Supporting Science 1 20:00 - 21:00 Virtual Tour of NIST Quantum Sensor Group Labs Wednesday 21 July 14:00 - 15:15 Oral O3: Instruments 15:15 - 15:25 Break 15:25 - 16:55 Oral O3: Instruments (continued) 16:55 - 17:05 Break 17:05 - 18:30 Poster P3: Instruments, Astrophysics and Cosmology 1 18:00 - 19:00 Vendor Exhibitor Hour Thursday 22 July 14:00 - 15:15 Oral O4A: Rare Events 1 Oral O4B: Material Analysis, Metrology, Other 15:15 - 15:25 Break 15:25 - 16:55 Oral O4A: Rare Events 1 (continued) Oral O4B: Material Analysis, Metrology, Other (continued) 16:55 - 17:05 Break 17:05 - 18:30 Poster P4: Rare Events, Materials Analysis, Metrology, Other Applications 20:00 - 21:00 Virtual Tour of NIST Cleanoom Monday 26 July 23:00 - 00:15 Oral O5: Devices 2 00:15 - 00:25 Break 00:25 - 01:55 Oral O5: Devices 2 (continued) 01:55 - 02:05 Break 02:05 - 03:30 Poster P5: MMCs, SNSPDs, more TESs Tuesday 27 July 23:00 - 00:15 Oral O6: Warm Readout and Supporting Science 00:15 - 00:25 Break 00:25 - 01:55 Oral O6: Warm Readout and Supporting Science -

Profile of Gary Ruvkun
PROFILE Profile of Gary Ruvkun wash in the faint glow of a fluo- Brush with Molecular Biology rescent lamp, a pair of serpentine The story of Ruvkun’s metamorphosis Anematode worms lie on a Petri from a keen undergraduate into a leading plate, their see-through bodies light in his field of study begins at Har- magnified 100-fold by one of several vard University, where he enrolled in microscopes arrayed in a darkened bay in a Ph.D. program in 1976 upon returning National Academy of Sciences member to the United States. Like many other Gary Ruvkun’s laboratory at Massachu- scientific institutions across the world in setts General Hospital. While one of the the mid-1970s, Harvard was astir with the worms wiggles its way around the plate, promise of recombinant DNA technol- the other shows no signs of life, ogy, and Ruvkun wasted no time em- its midsection ruptured and its innards bracing its tools. “My undergraduate strewn asunder. A filter slides into place, education had not prepared me at all for and the worms are bathed in a dull recombinant DNA, but I immersed my- green haze. The wiggling worm has a bea- self into its culture at Harvard, much of con of nerve cells in its head, the ganglia which was James Watson’s creation from lit up by a genetic trick that has rescued a decade earlier,” Ruvkun says. Propelled the worm from death; its neighbor wears Gary Ruvkun. by a desire to be a part of the culture of no such beacon. The worms were deprived basic molecular biology, all while per- of a tiny RNA molecule, called a micro- forming science with the potential to im- RNA, which helps shepherd them through not 5-year-old children. -

2008 Harvard / Paul F
The 2008 Harvard / Paul F. Glenn Symposium on Aging June 23, 2008 Paul F. Glenn Laboratories for the Biological Mechanisms of Aging Welcome to the 3rd Annual Harvard/Paul F. Glenn Symposium on Aging. Each year, the Paul F. Glenn Laboratories host the Harvard Symposium on Aging with a mission to educate the wider research community about advancements in this fast-paced field and to stimulate collaborative research in this area. We have been fortunate to have many of the leaders in the aging field speak at these symposia. As a result, attendees come not only from the Harvard research community but from across the nation and from overseas for this one day event. We are glad you could join us here today. The reasons for accelerating research molecular biology of aging are clear. First and foremost, the number of aged individuals in developed countries is growing rapidly, which is going to place an unprecedented burden on the families and the economies of those nations. Because chronic illness in the elderly is a major medical cost, enormous savings would be achieved if mortality and morbidity could be compressed within a shorter duration of time at the end of life. A study by the RAND Corporation in 2006 concluded that advances in medicine arising from aging research would be 10-100 times more cost-effective than any other medical breakthrough. Advances in aging research have shown that it is possible to extend the healthy lifespan of laboratory animals through genetic and pharmacological means. Many leaders in the aging field predict that significant strides will be made in understanding how human health and lifespan are regulated, leading to novel medicines to forestall and treat diseases of aging such as diabetes, cancer, Alzheimer’s and heart disease. -

Estimating the Overdispersion in COVID
Wellcome Open Research 2020, 5:67 Last updated: 13 JUL 2020 RESEARCH ARTICLE Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China [version 3; peer review: 2 approved] Akira Endo 1,2, Centre for the Mathematical Modelling of Infectious Diseases COVID-19 Working Group, Sam Abbott 1,3, Adam J. Kucharski1,3, Sebastian Funk 1,3 1Department of Infectious Disease Epidemiology, London School of Hygiene & Tropical Medicine, London, WC1E 7HT, UK 2The Alan Turing Institute, London, NW1 2DB, UK 3Centre for the Mathematical Modelling of Infectious Diseases, London School of Hygiene & Tropical Medicine, London, WC1E 7HT, UK First published: 09 Apr 2020, 5:67 Open Peer Review v3 https://doi.org/10.12688/wellcomeopenres.15842.1 Second version: 03 Jul 2020, 5:67 https://doi.org/10.12688/wellcomeopenres.15842.2 Reviewer Status Latest published: 10 Jul 2020, 5:67 https://doi.org/10.12688/wellcomeopenres.15842.3 Invited Reviewers 1 2 Abstract Background: A novel coronavirus disease (COVID-19) outbreak has now version 3 spread to a number of countries worldwide. While sustained transmission (revision) report chains of human-to-human transmission suggest high basic reproduction 10 Jul 2020 number R0, variation in the number of secondary transmissions (often characterised by so-called superspreading events) may be large as some countries have observed fewer local transmissions than others. version 2 Methods: We quantified individual-level variation in COVID-19 (revision) report report transmission by applying a mathematical model to observed outbreak sizes 03 Jul 2020 in affected countries. We extracted the number of imported and local cases in the affected countries from the World Health Organization situation report and applied a branching process model where the number of secondary version 1 transmissions was assumed to follow a negative-binomial distribution. -

Medical Innovation: Statins (Pharmaceutical: Small Molecule)
MEDICAL INNOVATION: STATINS (PHARMACEUTICAL: SMALL MOLECULE) Physicians: Akira Endo, biochemist who discovered the first statin, and Scott Grundy and David Bilheimer, lead investigators in clinical trials Industry: Sankyo, Merck & Co., Inc Situation A heart attack every 34 seconds Coronary artery disease -- the narrowing of blood vessels that supply the heart with oxygen – affects 16.8 million Americans and stands as the leading cause of death in the United States. The American Heart Association (AHA) estimates that about every 34 seconds, an American will have a heart attack. In addition, the lifetime risk of having cardiovascular disease after age 40 is 2 in 3 men and more than 1 in 2 women. One of the primary risk factors for coronary artery disease is high levels of LDL cholesterol, known as "bad" cholesterol because it can build up on the inside of arteries, causing them to become narrow from plaque. Until recently, people with high levels of LDL cholesterol had very few tools to lower this risk factor directly, and had to rely solely on radical diet modifications and other indirect means of combating this condition. Physician-Industry Collaboration Building on an earlier discovery In 1971, Akira Endo, a Japanese biochemist working for the drug company Sankyo, began the search for a cholesterol-lowering drug. Earlier research had already shown that the body manufactures cholesterol mostly in the liver, using a special enzyme. Endo and his team reasoned that certain microorganisms may produce inhibitors of this enzyme to defend themselves against other organisms. The first agent they identified was mevastatin, a molecule produced by the fungus Penicillium citrinum. -

Dr. Paul Janssen Award for Biomedical Research Issues 2015
Press Contacts: Dr. Paul Janssen Award for Biomedical Research Issues Seema Kumar 2015 Call for Nominations 908-405-1144 (M) [email protected] New Brunswick, N.J. – January 21, 2015 – The Dr. Paul Janssen Award for Diane Pressman Biomedical Research today opens its 2015 call for nominations. This prestigious 908-927-6171 (O) award recognizes individuals whose scientific research has made, or has the [email protected] potential to make, significant contributions toward the improvement of human Frederik Wittock health. Nominations will be accepted until March 15, 2015 at +32 14 60 57 24 (O) www.pauljanssenaward.com for consideration by an independent selection [email protected] committee of world renowned scientists. Beginning in 2015, the cash prize awarded to the scientist or group of scientists receiving the Award will be increased to $200,000. This increase in the monetary award reflects the growing importance of basic biomedical research, and continued recognition by Johnson & Johnson of excellence in the field. The Dr. Paul Janssen Award for Biomedical Research honors Dr. Paul Janssen (1926-2003), who is widely recognized as one of the most productive scientists of the 20th century. Known throughout the scientific community as “Dr. Paul,” Janssen was responsible for breakthrough treatments in disease areas including pain management, psychiatry, infectious disease and gastroenterology, and founded Janssen Pharmaceutica, N.V., a Johnson & Johnson Company. “Innovative science and technology have the power to transform the world,” said Paul Stoffels, M.D., Chief Scientific Officer and Worldwide Chairman, Pharmaceuticals, Johnson & Johnson. “Through the Dr. Paul Janssen Award for Biomedical Research, Johnson & Johnson honors the inspirational legacy of Dr. -

Statins: in the Beginning
J R Coll Physicians Edinb 2009; 39:362–4 Paper doi:10.4997/JRCPE.2009.425 © 2009 Royal College of Physicians of Edinburgh Statins: in the beginning 1KS Lyons; 2M Harbinson 1Research Fellow, Department of Therapeutics and Pharmacology, Queen’s University, Belfast; 2Senior Lecturer, Department of Therapeutics and Pharmacology, Queen’s University, Belfast, and Consultant Cardiologist, Belfast Health and Social Care Trust, UK ABSTRACT Since the first human trial of a hydroxymethylglutaryl-coenzyme A Published online December 2009 (HMG CoA) reductase inhibitor in 1978, the growth in importance of this drug class, both financially and medically, has been staggering. The aim of this paper is to Correspondence to KS Lyons, Department of Therapeutics and summarise how this drug class was developed, highlighting the role of Akira Endo. Pharmacology, Whitla Medical Building, 97 Lisburn Road, KEYWORDS Compactin, Akira Endo, lovastatin, statins Belfast BT9 7BL DECLARATION OF INTERESTS No conflict of interests declared. tel.+44 (0)2890 975772 e-mail [email protected] Hydroxymethylglutaryl-coenzyme A (HMG CoA) two years at the Albert Einstein College of Medicine reductase inhibitors (statins) are now one of the most working on cell wall biosynthetic enzymes.6 widely prescribed medications worldwide, with an estimated 2 million people in the UK1 and more than 18 When Endo returned to Sankyo in 1968 he began work million people in the USA currently taking a statin.2 on the quest to find an effective cholesterol-lowering From their development in the 1970s the impact of drug. He worked on the premise that some microbes statins on the treatment of cardiovascular disease been might produce inhibitors of enzymes necessary for staggering. -

Pjapan PRIZE NEWS=No.35=Ł\
THE SCIENCE AND TECHNOLOGY No. 35 FOUNDATION OF JAPAN (JSTF) January 2006 Akasaka Twin Tower East, 13th Floor, 17-22 Akasaka 2-chome, Minato-ku, Tokyo, 107-0052, JAPAN Tel. +81-3-5545-0551 Fax. +81-3-5545-0554 E-mail [email protected] URL http://www.japanprize.jp U.K. and Japanese Scientists Named as Laureates of the 2006(22nd)Japan Prize The Science and Technology Foundation of Japan (Chairman: Hiroyuki Yoshikawa) announced that British and Japanese scientists have been named as laureates of the 2006 (22nd) Japan Prize. Sir John Houghton (74), Formerly Chief Executive, Hadley Centre for Climate Prediction and Research, Meteorological Office, U.K., will receive the Japan Prize in this year's category of "Global Change." He contributed to the Atmospheric structure and composition based on his satel- ite observation technology and for promotion of international assessments of climate change. Dr. Akira Endo (72), Director, Biopharm Research Laboratories, Inc., Japan, will receive the Ja- pan Prize in the category of "The Development of Novel Therapeutic Concepts and Technolo- gies." He contributed to the Discovery of the Statins. The Development of Novel Global Change Therapeutic Concepts and Technologies Sir John Houghton CBE FRS Dr. Akira Endo Japan Prize The Japan Prize is awarded to world-class scientists and technolo- Fields of study for the prize encompass all categories of science gists who were credited with original and outstanding achievements and technology, with two categories designated for the prize each and contributed to the advancement of science and technology, year in consideration of developments in science and technology. -

Exploitation of Aspergillus Terreus for the Production of Natural Statins
Journal of Fungi Review Exploitation of Aspergillus terreus for the Production of Natural Statins Mishal Subhan 1, Rani Faryal 1 and Ian Macreadie 2,* 1 Department of Microbiology, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad 45320, Pakistan; [email protected] (M.S.); [email protected] (R.F.) 2 School of Science, RMIT University, Bundoora, Victoria 3083, Australia * Correspondence: [email protected]; Tel.: +61-3-9925-6627 Academic Editor: David S. Perlin Received: 19 April 2016; Accepted: 26 April 2016; Published: 30 April 2016 Abstract: The fungus Aspergillus (A.) terreus has dominated the biological production of the “blockbuster” drugs known as statins. The statins are a class of drugs that inhibit HMG-CoA reductase and lead to lower cholesterol production. The statins were initially discovered in fungi and for many years fungi were the sole source for the statins. At present, novel chemically synthesised statins are produced as inspired by the naturally occurring statin molecules. The isolation of the natural statins, compactin, mevastatin and lovastatin from A. terreus represents one of the great achievements of industrial microbiology. Here we review the discovery of statins, along with strategies that have been applied to scale up their production by A. terreus strains. The strategies encompass many of the techniques available in industrial microbiology and include the optimization of media and fermentation conditions, the improvement of strains through classical mutagenesis, induced genetic manipulation and the use of statistical design. Keywords: Aspergillus terreus; compactin; fermentation; industrial microbiology; lovastatin; mevastatin; mutagenesis; optimization; polyketide 1. Introduction Statins are polyketide compounds that are produced by some fungi during their secondary metabolism [1]. -

Table of Contents (PDF)
July 5, 2016 u vol. 113 u no. 27 From the Cover 7308 Finding causality in Big Data E3901 Treating inflammation-driven fibrosis 7403 Interactions between liquid droplets on solids 7667 Role of plant photoreceptors Contents THIS WEEK IN PNAS 7285 In This Issue Cover image: Societies now collect immense amounts of data from both LETTERS (ONLINE ONLY) controlled experiments and natural E3811 Avoid the hard problem: Employment of mental simulation for prediction is observations such as activity on the already a crucial step World Wide Web. Pictured is a Malte Schilling and Holk Cruse visualization of one possible archive of E3812 Consciousness explained or consciousness redefined? data in network form. Such databases Shelley Anne Adamo are analyzed and used for scientific, business, and health purposes. Effective E3813 Insects cannot tell us anything about subjective experience or the origin use of these large databases of of consciousness information, collectively termed Big Brian Key, Robert Arlinghaus, and Howard I. Browman Data, requires identifying the causal E3814 Reply to Adamo, Key et al., and Schilling and Cruse: Crawling around the hard forces behind patterns in the data. Such problem of consciousness an effort was the focus of the Sackler Colin Klein and Andrew B. Barron Colloquium on Drawing Causal Inference from Big Data. See the OPINION—Leading scientists discuss current issues Introduction to the Colloquium papers by Richard M. Shiffrin on pages 7308– 7287 In the wake of Paris Agreement, scientists must embrace new directions -

By Sean B. Carroll
Educator Supplements to the book, THE SERENGETI RULES THE QUEST TO DISCOVER HOW LIFE WORKS AND WHY IT MATTERS BY SEAN B. CARROLL Prepared by Paul Strode Teacher of Biology at Fairview High School Boulder, Colorado 1 TABLE OF CONTENTS I. SUMMARY OF THE SERENGETI RULES 4–8 II. INTRODUCTION TO QUESTION SETS FOR THE SERENGETI RULES 9–11 III. ENGAGEMENT QUESTIONS 12–21 IV. DISCUSSION QUESTIONS 22–30 V. ASSESSMENT QUESTIONS 31–40 VI. COURSE CURRICULUM ALIGNMENTS: AP BIOLOGY, IB BIOLOGY, NGSS 41–53 VII. EXAMPLE ADVANCED BIOLOGY CURRICULUM MAP FOR THE SERENGETI RULES 54–57 2 HOW TEACHERS MIGHT USE THE SERENGETI RULES IN THE CLASSROOM The Serengeti Rules, molecular biologist Sean Carroll takes the reader on a journey through the history of the scientific discovery of many of nature’s most important regulatory mechanisms, from molecules to megafauna. In the spirit of Theodisius Dobzhansky, Carroll teaches us in The Serengeti Rules that nothing in biology makes sense except in the light of regulation. Thus, at the very center of the book is the big, biological idea of homeostasis. Indeed, a key to understand- ing how living systems work and the supporting roles played by evolutionary processes is understanding how a rela- tively stable environment, be it in a cell or in an ecosystem, is maintained and regulated. Throughout the book, Carroll tells the stories of the scientists—and their discoveries—that have given us our current understanding of regulation in nature. But in the stories, Carroll also teaches the reader about the nature of science and scientific reasoning, the impor- tance of modeling, and how scientific discovery is full of determination and success but also frequented with failure.