Ncomms15700.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

P2Y6 Receptors Regulate CXCL10 Expression and Secretion in Mouse Intestinal Epithelial Cells
fphar-09-00149 February 26, 2018 Time: 17:57 # 1 ORIGINAL RESEARCH published: 28 February 2018 doi: 10.3389/fphar.2018.00149 P2Y6 Receptors Regulate CXCL10 Expression and Secretion in Mouse Intestinal Epithelial Cells Mabrouka Salem1,2, Alain Tremblay2, Julie Pelletier2, Bernard Robaye3 and Jean Sévigny1,2* 1 Département de Microbiologie-Infectiologie et d’Immunologie, Faculté de Médecine, Université Laval, Québec City, QC, Canada, 2 Centre de Recherche du CHU de Québec – Université Laval, Québec City, QC, Canada, 3 Institut de Recherche Interdisciplinaire en Biologie Humaine et Moléculaire, Université Libre de Bruxelles, Gosselies, Belgium In this study, we investigated the role of extracellular nucleotides in chemokine (KC, MIP- 2, MCP-1, and CXCL10) expression and secretion by murine primary intestinal epithelial cells (IECs) with a focus on P2Y6 receptors. qRT-PCR experiments showed that P2Y6 was the dominant nucleotide receptor expressed in mouse IEC. In addition, the P2Y6 Edited by: ligand UDP induced expression and secretion of CXCL10. For the other studies, we Kenneth A. Jacobson, −=− National Institutes of Health (NIH), took advantage of mice deficient in P2Y6 (P2ry6 ). Similar expression levels of P2Y1, −=− United States P2Y2, P2X2, P2X4, and A2A were detected in P2ry6 and WT IEC. Agonists of Reviewed by: TLR3 (poly(I:C)), TLR4 (LPS), P2Y1, and P2Y2 increased the expression and secretion Fernando Ochoa-Cortes, of CXCL10 more prominently in P2ry6−=− IEC than in WT IEC. CXCL10 expression Universidad Autónoma de San Luis −=− Potosí, Mexico and secretion induced by poly(I:C) in both P2ry6 and WT IEC were inhibited by Markus Neurath, general P2 antagonists (suramin and Reactive-Blue-2), by apyrase, and by specific Universitätsklinikum Erlangen, Germany antagonists of P2Y1, P2Y2, P2Y6 (only in WT), and P2X4. -

Chemokine Receptors in Allergic Diseases Laure Castan, A
Chemokine receptors in allergic diseases Laure Castan, A. Magnan, Grégory Bouchaud To cite this version: Laure Castan, A. Magnan, Grégory Bouchaud. Chemokine receptors in allergic diseases. Allergy, Wiley, 2017, 72 (5), pp.682-690. 10.1111/all.13089. hal-01602523 HAL Id: hal-01602523 https://hal.archives-ouvertes.fr/hal-01602523 Submitted on 11 Jul 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - ShareAlike| 4.0 International License Allergy REVIEW ARTICLE Chemokine receptors in allergic diseases L. Castan1,2,3,4, A. Magnan2,3,5 & G. Bouchaud1 1INRA, UR1268 BIA; 2INSERM, UMR1087, lnstitut du thorax; 3CNRS, UMR6291; 4Universite de Nantes; 5CHU de Nantes, Service de Pneumologie, Institut du thorax, Nantes, France To cite this article: Castan L, Magnan A, Bouchaud G. Chemokine receptors in allergic diseases. Allergy 2017; 72: 682–690. Keywords Abstract asthma; atopic dermatitis; chemokine; Under homeostatic conditions, as well as in various diseases, leukocyte migration chemokine receptor; food allergy. is a crucial issue for the immune system that is mainly organized through the acti- Correspondence vation of bone marrow-derived cells in various tissues. Immune cell trafficking is Gregory Bouchaud, INRA, UR1268 BIA, rue orchestrated by a family of small proteins called chemokines. -

Immunosuppression Via Adenosine Receptor Activation by Adenosine
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Crossref RESEARCH ARTICLE elifesciences.org Immunosuppression via adenosine receptor activation by adenosine monophosphate released from apoptotic cells Hiroshi Yamaguchi1, Toshihiko Maruyama2, Yoshihiro Urade2†, Shigekazu Nagata1,3* 1Department of Medical Chemistry, Graduate School of Medicine, Kyoto University, Kyoto, Japan; 2Department of Molecular Behavioral Biology, Osaka Bioscience Institute, Osaka, Japan; 3Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation, Kyoto, Japan Abstract Apoptosis is coupled with recruitment of macrophages for engulfment of dead cells, and with compensatory proliferation of neighboring cells. Yet, this death process is silent, and it does not cause inflammation. The molecular mechanisms underlying anti-inflammatory nature of the apoptotic process remains poorly understood. In this study, we found that the culture supernatant of apoptotic cells activated the macrophages to express anti-inflammatory genes such as Nr4a and Thbs1. A high level of AMP accumulated in the apoptotic cell supernatant in a Pannexin1-dependent manner. A nucleotidase inhibitor and A2a adenosine receptor antagonist inhibited the apoptotic *For correspondence: snagata@ supernatant-induced gene expression, suggesting AMP was metabolized to adenosine by an mfour.med.kyoto-u.ac.jp ecto-5’-nucleotidase expressed on macrophages, to activate the macrophage A2a adenosine receptor. Intraperitoneal injection of zymosan into Adora2a- or Panx1-deficient mice produced † Present address: Molecular high, sustained levels of inflammatory mediators in the peritoneal lavage. These results indicated Sleep Biology Laboratory, that AMP from apoptotic cells suppresses inflammation as a ‘calm down’ signal. WPI-International Institute for DOI: 10.7554/eLife.02172.001 Integrative Sleep Medicine, University of Tsukuba, Ibaraki, Japan Competing interests: The Introduction authors declare that no competing interests exist. -
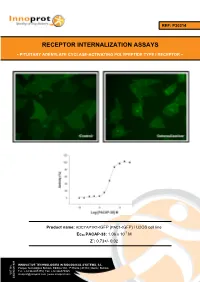
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Atypical Chemokine Receptor 4 Shapes Activated B Cell Fate
Brief Definitive Report Atypical chemokine receptor 4 shapes activated B cell fate Ervin E. Kara,1 Cameron R. Bastow,1 Duncan R. McKenzie,1 Carly E. Gregor,1 Kevin A. Fenix,1 Rachelle Babb,1 Todd S. Norton,1 Dimitra Zotos,3 Lauren B. Rodda,4 Jana R. Hermes,6 Katherine Bourne,6 Derek S. Gilchrist,7 Robert J. Nibbs,7 Mohammed Alsharifi,1 Carola G. Vinuesa,8 David M. Tarlinton,3,9 Robert Brink,6,10 Geoffrey R. Hill,11 Jason G. Cyster,4,5 Iain Comerford,1 and Shaun R. McColl1,2 1Department of Molecular and Cellular Biology, School of Biological Sciences and 2Centre for Molecular Pathology, School of Biological Sciences, University of Adelaide, Adelaide, South Australia, Australia 3Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia 4Department of Microbiology and Immunology and 5Howard Hughes Medical Institute, Department of Microbiology and Immunology, University of California, Downloaded from http://rupress.org/jem/article-pdf/215/3/801/1168927/jem_20171067.pdf by guest on 28 September 2021 San Francisco, San Francisco, CA 6Immunology Division, Garvan Institute of Medical Research, Darlinghurst, New South Wales, Australia 7Institute of Infection, Immunity and Inflammation, College of Medicine, Veterinary and Life Sciences, University of Glasgow, Glasgow, Scotland, UK 8Department of Immunology and Infectious Disease, John Curtin School of Medical Research, Australian National University, Canberra, Australian Capital Territory, Australia 9Department of Immunology and Pathology, Monash University, Melbourne, Victoria, Australia 10St Vincent’s Clinical School, University of New South Wales, Darlinghurst, New South Wales, Australia 11Immunology Department, QIMR Berghofer Medical Research Institute, Brisbane, Queensland, Australia Activated B cells can initially differentiate into three functionally distinct fates—early plasmablasts (PBs), germinal center (GC) B cells, or early memory B cells—by mechanisms that remain poorly understood. -

Atypical Chemokine Receptors and Their Roles in the Resolution of the Inflammatory Response
REVIEW published: 10 June 2016 doi: 10.3389/fimmu.2016.00224 Atypical Chemokine Receptors and Their Roles in the Resolution of the inflammatory Response Raffaella Bonecchi1,2 and Gerard J. Graham3* 1 Humanitas Clinical and Research Center, Rozzano, Italy, 2 Department of Biomedical Sciences, Humanitas University, Rozzano, Italy, 3 Chemokine Research Group, Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow, UK Chemokines and their receptors are key mediators of the inflammatory process regulating leukocyte extravasation and directional migration into inflamed and infected tissues. The control of chemokine availability within inflamed tissues is necessary to attain a resolving environment and when this fails chronic inflammation ensues. Accordingly, vertebrates have adopted a number of mechanisms for removing chemokines from inflamed sites to help precipitate resolution. Over the past 15 years, it has become apparent that essential players in this process are the members of the atypical chemokine receptor (ACKR) family. Broadly speaking, this family is expressed on stromal cell types and scavenges Edited by: Mariagrazia Uguccioni, chemokines to either limit their spatial availability or to remove them from in vivo sites. Institute for Research in Biomedicine, Here, we provide a brief review of these ACKRs and discuss their involvement in the Switzerland resolution of inflammatory responses and the therapeutic implications of our current Reviewed by: knowledge. Mette M. M. Rosenkilde, University of Copenhagen, Keywords: chemokines, immunity, inflammation, scavenging, atypical receptors Denmark Mario Mellado, Spanish National Research Council, Spain INTRODUCTION *Correspondence: Gerard J. Graham An effective inflammatory response requires carefully regulated initiation, maintenance, and [email protected] resolution phases (1). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

In This Issue
THE JOURNAL OF IMMUNOLOGY IN THIS ISSUE Limiting C damage rather than decreased proliferation after recruitment by measur- ing infiltrating donor cells 6 h after injection. CXCR6 defi- ϩ reventing the forma- ciency did not alter levels of donor CD8 T cells in the blood or tion of the C3 conver- spleen compared with wild-type donor cells. The data support a ϩ P tase reduces damage specific role for CXCR6 in homing of activated donor CD8 T from inflammation following cells to the inflamed liver in graft-vs-host-induced hepatitis. activation of the mammalian C classical pathway. Schisto- soma complement C2 receptor inhibitor trispanning (CRIT) TLR enhancement of diabetes is known to bind C2 and pre- vent its cleavage. Inal et al. (p. 356) found CRIT homologues in ilham rat virus (KRV) infection of BBDR rats results genomic DNA from Trypanosoma, human, rat, and cod by in type 1 diabetes in ϳ25% of the animals. The fre- Southern blotting and PCR. Ab against an extracellular N- K quency is increased to 100% in rats pretreated with terminal peptide enabled detection of CRIT on the surface of the TLR3 agonist poly(I:C). To investigate the role of TLR- Jurkat cells by flow cytometry. CRIT expression in a wide range induced activation of innate immunity in autoimmune diabetes of human tissues and cells was shown using Abs against the induction, Zipris et al. (p. 131) found that pretreatment of N-terminal peptide and other regions of the protein. The bind- BBDR rats with other TLR ligands, including heat-killed bac- ing of biotinylated C2 to filter-immobilized CRIT or of fluo- teria, also enhanced KRV-induced diabetes induction; some li- rescence-labeled C2 to Jurkat cells was abolished by preincuba- gands were effective at low viral doses that were not usually di- tion of the filter or cells with anti-N-terminal peptide Ab; abetogenic. -

G Protein-Coupled Receptors As Therapeutic Targets for Multiple Sclerosis
npg GPCRs as therapeutic targets for MS Cell Research (2012) 22:1108-1128. 1108 © 2012 IBCB, SIBS, CAS All rights reserved 1001-0602/12 $ 32.00 npg REVIEW www.nature.com/cr G protein-coupled receptors as therapeutic targets for multiple sclerosis Changsheng Du1, Xin Xie1, 2 1Laboratory of Receptor-Based BioMedicine, Shanghai Key Laboratory of Signaling and Disease Research, School of Life Sci- ences and Technology, Tongji University, Shanghai 200092, China; 2State Key Laboratory of Drug Research, the National Center for Drug Screening, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 189 Guo Shou Jing Road, Pudong New District, Shanghai 201203, China G protein-coupled receptors (GPCRs) mediate most of our physiological responses to hormones, neurotransmit- ters and environmental stimulants. They are considered as the most successful therapeutic targets for a broad spec- trum of diseases. Multiple sclerosis (MS) is an inflammatory disease that is characterized by immune-mediated de- myelination and degeneration of the central nervous system (CNS). It is the leading cause of non-traumatic disability in young adults. Great progress has been made over the past few decades in understanding the pathogenesis of MS. Numerous data from animal and clinical studies indicate that many GPCRs are critically involved in various aspects of MS pathogenesis, including antigen presentation, cytokine production, T-cell differentiation, T-cell proliferation, T-cell invasion, etc. In this review, we summarize the recent findings regarding the expression or functional changes of GPCRs in MS patients or animal models, and the influences of GPCRs on disease severity upon genetic or phar- macological manipulations. -

BD Biosciences New RUO Reagents - November 2020
BD Biosciences New RUO reagents - November 2020 Reactivity Description Format Clone Size Cat. number Hu CD133 FITC W6B3C1 100µg 567029 Hu CD133 FITC W6B3C1 25µg 567033 Hu CD39 PE A1/CD39 100Tst 567156 Hu CD39 PE A1/CD39 25Tst 567157 Hu KIR2DL1/S1/S3/S5 PE HP-MA4 100Tst 567158 Hu KIR2DL1/S1/S3/S5 PE HP-MA4 25Tst 567159 Hu IL-22 Alexa Fluor® 647 MH22B2 100µg 567160 Hu IL-22 Alexa Fluor® 647 MH22B2 25µg 567161 Hu CD99 R718 TU12 50µg 751651 Hu CD161 R718 DX12 50µg 751652 Hu CD116 R718 HGMCSFR-M1 50µg 751653 Hu HLA-G R718 87G 50µg 751670 Hu CD27 R718 O323 50µg 751686 Hu CD80 (B7-1) R718 2D10.4 50µg 751737 Hu Integrin αvβ5 R718 ALULA 50µg 751738 Hu CD266 (Tweak-R) R718 ITEM-4 50µg 751739 Hu ErbB3 (HER-3) R718 SGP1 50µg 751799 Hu TCR Vβ5.1 R718 LC4 50µg 751816 Hu CD123 (IL-3Ra) R718 6H6 50µg 751844 Hu CD1a R718 SK9 50µg 751847 Hu CD20 R718 L27 50µg 751849 Hu Disial GD2 R718 14.G2A 50µg 751851 Reactivity Description Format Clone Size Cat. number Hu CD71 R718 L01.1 50µg 751853 Hu CD278 (ICOS) R718 DX29 50µg 751854 Hu B7-H4 R718 MIH43 50µg 751857 Hu CD53 R718 HI29 50µg 751858 Hu CD197 (CCR7) R718 2-L1-A 50µg 751859 Hu CD197 (CCR7) R718 3D12 50µg 751861 Hu CD31 R718 L133.1 50µg 751863 Hu EGF Receptor R718 EMAB-134 50µg 751864 Hu CD8b R718 2ST8.5H7 50µg 751867 Hu CD31 R718 MBC 78.2 50µg 751869 Hu CD162 R718 KPL-1 50µg 751873 Hu CD24 R718 ML5 50µg 751874 Hu CD159C (NKG2C) R718 134591 50µg 751876 Hu CD169 (Siglec-1) R718 7-239 50µg 751877 Hu CD16b R718 CLB-GRAN11.5 50µg 751880 Hu IgM R718 UCH-B1 50µg 751881 Hu CD275 R718 2D3/B7-H2 50µg 751883 Hu CD307e -

Profound Treg Perturbations Correlate with COVID-19 Severity
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.11.416180; this version posted December 15, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Profound Treg perturbations correlate with COVID-19 severity Silvia Galván-Peña1*, Juliette Leon1,2*, Kaitavjeet Chowdhary1, Daniel A. Michelson1, Brinda Vijaykumar1, Liang Yang1, Angela Magnuson1, Zachary Manickas-Hill3,4, Alicja Piechocka- Trocha3,4, Daniel P. Worrall3,4, Kathryn E. Hall3,5, Musie Ghebremichael3,4, Bruce D. Walker3,4,6, Jonathan Z. Li3,7, Xu G. Yu3,4, MGH COVID-19 Collection & Processing Team, Diane Mathis1 and Christophe Benoist1,8,+ 1Department of Immunology, Harvard Medical School, Boston, MA, USA 2 INSERM UMR 1163, University of Paris, Imagine Institute, Paris, France 3Massachusetts Consortium on Pathogen Readiness, Boston, MA, USA 4Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA 5Department of Medicine, Massachusetts General Hospital, Boston, MA, USA 6Howard Hughes Medical Institute, Center for the AIDS Programme of Research in South Africa. 7Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA 8Lead contact * Equal contribution +Address correspondence to: Christophe Benoist Department of Immunology Harvard Medical School 77 Avenue Louis Pasteur, Boston, MA 02115 e-mail: [email protected] Phone: (617) 432-7741 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.12.11.416180; this version posted December 15, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.