Clupeiformes’ Egg Envelope Proteins Characterization: the Case
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

European Anchovy Engraulis Encrasicolus (Linnaeus, 1758) From
European anchovy Engraulis encrasicolus (Linnaeus, 1758) from the Gulf of Annaba, east Algeria: age, growth, spawning period, condition factor and mortality Nadira Benchikh, Assia Diaf, Souad Ladaimia, Fatma Z. Bouhali, Amina Dahel, Abdallah B. Djebar Laboratory of Ecobiology of Marine and Littoral Environments, Department of Marine Science, Faculty of Science, University of Badji Mokhtar, Annaba, Algeria. Corresponding author: N. Benchikh, [email protected] Abstract. Age, growth, spawning period, condition factor and mortality were determined in the European anchovy Engraulis encrasicolus populated the Gulf of Annaba, east Algeria. The age structure of the total population is composed of 59.1% females, 33.5% males and 7.4% undetermined. The size frequency distribution method shows the existence of 4 cohorts with lengths ranging from 8.87 to 16.56 cm with a predominance of age group 3 which represents 69.73% followed by groups 4, 2 and 1 with respectively 19.73, 9.66 and 0.88%. The VONBIT software package allowed us to estimate the growth parameters: asymptotic length L∞ = 17.89 cm, growth rate K = 0.6 year-1 and t0 = -0.008. The theoretical maximum age or tmax is 4.92 years. The height-weight relationship shows that growth for the total population is a major allometry. Spawning takes place in May, with a gonado-somatic index (GSI) of 4.28% and an annual mean condition factor (K) of 0.72. The total mortality (Z), natural mortality (M) and fishing mortality (F) are 2.31, 0.56 and 1.75 year-1 respectively, with exploitation rate E = F/Z is 0.76 is higher than the optimal exploitation level of 0.5. -

R M , July1979 Rum, Jlilio1979
FA0 Fisheriee Circular No. 706 FIR/C706 FA0 Cimulaire mur lee p8ohes No 706 FAO, CirouLerem de Pom~fbNo 706 SELECTED BIBLIOUUPHY ON PELAGIC FISH EGG AND LARVA SURVEYS BIBLIOWHIE SELECTIVE SUR LES PROSPECTIONS D'OEUFS ET DE LAFNFS DE POISSONS PELAGIQUES BIBLIOWA SELECCIONADA SOBRE RECONOCIMIEN'IQS DE HUEVOS Y LARVAS DE PECES PEZAOICOS Prepared by/Prdparge par/Preparada por Paul E. Smith Southwest Fisheries Center La Jolla, California, U.S.A. t /Y Sally L. Richardem Oregon State University Corvallis, Oregon, U.S.A. FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS ORGANISATION DES NATIONS UNIES POUR L'ALIMZNTATIW ET L'AaRICUL'NRE ORC;BFIZACI(YN DE LAS NACI- UNIDAS PAR4 LA AaRICUL"RA Y LA ALIMENTACION Rm, July1979 Ram, juillet 1979 Rum, jlilio1979 -1- 1. SCOPE, COVERAGE AND ORGAMI~TION This bibliography is intended to provide aocass to published information on ichthyo- plankton survey methods, identification of fish -,and larvae, and results of meyn thet have been carried out in the put. Although the bibliograpb in selective, its coverage in- cludes all published works through 1973, and it htu been extended by the addition of all papers resented at the Oban Symposium and publinhed in the eJwposiun proceedings (Blazter, J.H.S. red.) 1974. The early life history of firh. SpringarcVerlsg, Berlin). The fint five seotiau of this bibliomp4y list worh by name and drte anly aooordinn to subject category. Section 2 containe refer.noe8 on survey equipment and mothods. Section 3 includes descriptions of early life rtages organized by taxonomic group. Section 4 lists references on species identification of fish aggm and larva4 by region (specifically, by FA0 etatietical area). -

Yellow Sea East China Sea
Yellow Sea East China Sea [59] 86587_p059_078.indd 59 1/19/05 9:18:41 PM highlights ■ The Yellow Sea / East China Sea show strong infl uence of various human activities such as fi shing, mariculture, waste discharge, dumping, and habitat destruction. ■ There is strong evidence of a gradual long-term increase in the sea surface temperature since the early 1900s. ■ Given the variety of forcing factors, complicated changes in the ecosystem are anticipated. ■ Rapid change and large fl uctuations in species composition and abundance in the major fi shery have occurred. Ocean and Climate Changes [60] 86587_p059_078.indd 60 1/19/05 9:18:48 PM background The Yellow Sea and East China Sea are epi-continental seas bounded by the Korean Peninsula, mainland China, Taiwan, and the Japanese islands of Ryukyu and Kyushu. The shelf region shallower than 200m occupies more The coasts of the Yellow Sea have diverse habitats due than 70% of the entire Yellow Sea and the East China Sea. to jagged coastlines and the many islands scattered The Yellow Sea is a shallow basin with a mean depth around the shallow sea. Intertidal fl at is the most of 44 m. Its area is about 404,000 km2 if the Bohai signifi cant coastal habitat. The tidal fl at in the Yellow Sea in the north is excluded. A trough with a maximum Sea consists of several different types such as mudfl at depth of 103 m lies in the center. Water exchange is with salt marsh, sand fl at with gravel beach, sand dune slow and residence time is estimated to be 5-6 years.37 or eelgrass bed, and mixed fl at. -

ASFIS ISSCAAP Fish List February 2007 Sorted on Scientific Name
ASFIS ISSCAAP Fish List Sorted on Scientific Name February 2007 Scientific name English Name French name Spanish Name Code Abalistes stellaris (Bloch & Schneider 1801) Starry triggerfish AJS Abbottina rivularis (Basilewsky 1855) Chinese false gudgeon ABB Ablabys binotatus (Peters 1855) Redskinfish ABW Ablennes hians (Valenciennes 1846) Flat needlefish Orphie plate Agujón sable BAF Aborichthys elongatus Hora 1921 ABE Abralia andamanika Goodrich 1898 BLK Abralia veranyi (Rüppell 1844) Verany's enope squid Encornet de Verany Enoploluria de Verany BLJ Abraliopsis pfefferi (Verany 1837) Pfeffer's enope squid Encornet de Pfeffer Enoploluria de Pfeffer BJF Abramis brama (Linnaeus 1758) Freshwater bream Brème d'eau douce Brema común FBM Abramis spp Freshwater breams nei Brèmes d'eau douce nca Bremas nep FBR Abramites eques (Steindachner 1878) ABQ Abudefduf luridus (Cuvier 1830) Canary damsel AUU Abudefduf saxatilis (Linnaeus 1758) Sergeant-major ABU Abyssobrotula galatheae Nielsen 1977 OAG Abyssocottus elochini Taliev 1955 AEZ Abythites lepidogenys (Smith & Radcliffe 1913) AHD Acanella spp Branched bamboo coral KQL Acanthacaris caeca (A. Milne Edwards 1881) Atlantic deep-sea lobster Langoustine arganelle Cigala de fondo NTK Acanthacaris tenuimana Bate 1888 Prickly deep-sea lobster Langoustine spinuleuse Cigala raspa NHI Acanthalburnus microlepis (De Filippi 1861) Blackbrow bleak AHL Acanthaphritis barbata (Okamura & Kishida 1963) NHT Acantharchus pomotis (Baird 1855) Mud sunfish AKP Acanthaxius caespitosa (Squires 1979) Deepwater mud lobster Langouste -

Online First Article
Pakistan J. Zool., pp 1-10, 2021. DOI: https://dx.doi.org/10.17582/journal.pjz/20200222110209 Preliminary Assessment of a Coupled Dynamic-Energy Budget and Agent-based Model (DEB-ABM) for Predicting Individual and Population-Level Dynamics: A Case Study on Anchovy, Engraulis japonicus Baochao Liao1,2, Xiujuan Shan2,3* and Yunlong Chen2,3 1Department of Mathematics and Statistics, Shandong University, Weihai, Shandong 264209, China 2Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, Shandong 266237, China 3 Article Information Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Received 22 February 2020 Qingdao, Shandong 266071, China Revised 04 April 2020 Accepted 23 April 2020 Available online 04 September 2020 ABSTRACT Authors’ Contribution Dynamic energy budget (DEB; also known as Kooijman–Metz DEB) theory is a well-tested framework BL conceived and designed the study. for modelling energy acquisition, and for describing vital rates at which organisms acquire and use energy, XS arranged the data. BL and YC such as for growth and reproduction. The coupling of a DEB with an agent-based model (generating helped in data analysis. YC prepared the Figures. BL analyzed the data and a DEB- ABM) enables examination of the effects of environmental change at a population-level on a wrote the article. species to be examined. The present study applied a DEB-ABM to the Japanese anchovy Engraulis japonicus. The DEB-ABM accurately captured energy acquisition and allocation throughout the anchovy Key words lifecycle (egg, yolk sac larva, exogenous feeding larva, juvenile, and adult) and predicted how individual- Dynamic energy budget, Agent-based level processes affect energy dynamics at higher levels of biological organization. -

Differences in Foods and Feeding Habits in Common Minke and Sei Whales in the Western North Pacific Based on Samples Under JARPN II Survey Project
Differences in foods and feeding habits in common minke and sei whales in the western North Pacific based on samples under JARPN II survey project Ryosuke Okamoto1, Tsutomu Tamura2, Kenji Konishi2 and Hidehiro Kato2 1 Tokyo University of Marine Science and Technology, Japan 2 The Institute of Cetacean Research, Japan Aim of this Study 1.To clarify geographical and monthly changes in prey composition 2.To clarify changes prey prey composition with growth 3.To compare prey among whale species Feeding habits of minke and sei whale in the PICES area in conjunction with temporal and spatial variation 2006 Common minke whale Balaenoptera acutorostrata Body length : 8 m Body weight : 6 t Sei whale Balaenoptera borealis Body length : 14.5 m Body weight : 16 t Materials and methods The Second Phase of Japanese Whale Research Program under Special Permit in the western North Pacific (JARPNⅡ) Sampling location of common minke and sei whale Common minke whale 50°N Sei whale JARPN Ⅱ 2006 ● May to August ● From the Pacific coast of Japan to 170°E and north of 35N 40°N Western Central Eastern Sub-areas 140°E 150°E 160°E 170°E Sampling of stomach contents Stomach contents from each forestomach were weighed to the nearest 0.1 kg. A part of each stomach contents (sub-sample) was taken, weight and frozen or fixed in 10% formalin solution (for crustacean) for later analyses. Anchovies from a forestomach of sei whale Estimation of amount eaten by whales Identification of species Japanese anchovy Otolith (Japanese anchovy) Number of individuals for OL=4.1mm -

Supplemental Material Evaluation of the Global Impacts of Mitigation on Persistent, Bioaccumulative and Toxic Pollutants in Mari
Supplemental Material Evaluation of the global impacts of mitigation on persistent, bioaccumulative and toxic pollutants in marine fish. Lindsay T. Bonito, Amro Hamdoun, Stuart A. Sandin Marine Biology Research Department, Scripps Institution of Oceanography, 9500 Gilman Drive, La Jolla, CA 92093-0202, USA Table of Contents Supplemental Figure 1: Regional Data Distribution 2 …………………………………………………………… Supplemental Figure 2: Habitat Data Distribution .. 3 …………………………………………………… ……… Supplemental Figure 3: Regional Temporal Analysis.. .. 4 ……………………………………… ……… ……… Supplemental Table 1: Sample Sizes and Data Distribution ... .. ... 5 ………………………………… … …… … Supplemental Table 2: ANOVA Summary Table (Figure 2) .. .. 6 … ………………………………… … …… … Supplemental Table 3: ANOVA Summary Table (Figure 3) .. ... .. .. 6 … ……………………………… … …… … Supplemental Table 4: ANOVA Summary Table (Figure 4) .. .. 7 … ………………………………… … …… … Supplemental Table 5: Linear Regression Summary (Figure 5).. .. .. 7 ……………………………… … ……… Supplemental Table 6: Linear Regression Summary, Years 1990-2012. ... .... .. 7 … ……………… ……… … Supplemental Table 7: Species List .. ... .. 8 …………………………………………… ……………… ……… … … Supplemental Table 8: Seafood Database Reference List . 26 ………………………… ………… …… ……… 1 Supplemental Figure 1: Regional Data Distribution. Data distribution across pollutant groups, regions, and decades. Size of pie chart reflects number of data points included in analysis for each region. 5 global regions aggregated: EPO East Pacific Ocean; WPO West Pacific Ocean; -

Morphological Divergence in the Anchovy Anchoa Januaria
Journal of the Marine Morphological divergence in the anchovy Biological Association of the United Kingdom Anchoa januaria (Actinopterygii, Engraulidae) between tropical and subtropical estuarine cambridge.org/mbi areas on the Brazilian coast Joaquim N. S. Santos1,2, Rafaela de S. Gomes-Gonçalves1, Márcio de A. Silva3, Original Article Ruan M. Vasconcellos4 and Francisco G. Araújo1 Cite this article: Santos JNS, Gomes- 1Laboratório de Ecologia de Peixes, Universidade Federal Rural do Rio de Janeiro, BR 465, km 7, Seropédica, RJ, Gonçalves R de S, Silva M de A, Vasconcellos 2 RM, Araújo FG (2019). Morphological CEP 23890-000, Brazil; Instituto Federal do Norte de Minas Gerais, Campus Almenara, Rodovia BR 367, Km 07, s/n 3 divergence in the anchovy Anchoa januaria – Zona Rural, Almenara, MG, CEP 39900-000, Brazil; Agência Nacional de Águas, Superintendência de (Actinopterygii, Engraulidae) between tropical Planejamento em Recursos Hídricos, Setor Policial Sul, Área 5, Quadra 3, Bloco L, Sala 218, 70610-200, Brasilia, DF, and subtropical estuarine areas on the Brazil and 4Instituto Federal de Educação, Ciência e Tecnologia de Mato Grosso do Sul, Campus Ponta Porã, BR Brazilian coast. Journal of the Marine Biological 463, Km 14, Sanga Puitã, Ponta Porã, MS, Caixa Postal 287, Brazil Association of the United Kingdom 99,947–955. https://doi.org/10.1017/S0025315418000802 Abstract Received: 13 July 2018 Phenotypic differentiation among fish populations may be used for management of distinct Revised: 23 August 2018 stocks and helps in conserving biodiversity. We compared morphometric and meristic char- Accepted: 24 August 2018 First published online: 8 October 2018 acters of the anchovy Anchoa januaria from shallow semi-closed bays between the south-east- ern (Tropical, 23°S) and southern (Subtropical, 25°S) Brazilian coast. -
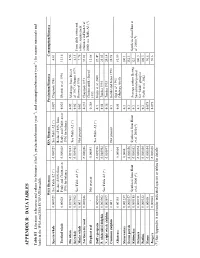
Appendix B: Da T a T a Bles
APPENDIX B: DATA TABLES Table B1 Literature references and values for biomass (t/km2), production/biomass (year-1), and consumption/biomass (year-1) for marine mammals and birds in the WSA and ESA ECOPATH models. WSA Biomass ESA Biomass Production/Biomass Consumption/Biomass Sperm whale 0.000929 See Table A2 (*) 0.000929 See Table A2 (*) 0.059 Chapman 1980 6.61 Reduced 80% from Reduced 80% from Toothed whale 0.00028 Pauly and Christensen 0.000028 Pauly and Christensen 0.025 Olesiuk et al. 1990 11.16 1996 for balance 1996 for balance Fin whale 0.027883 0.027883 0.02 4.56 See Table A2 (*) Midrange from Doi et Sei whale 0.005902 0.005902 0.02 al. 1967, Ohsumi 1979, 6.15 From daily ration and See Table A2 (*) Horwood 1990 Minke whale 0.001 - Not present 0.02 7.78 caloric requirement Northern fur seal 0.000246 0.000246 0.235 Chapman 1973 39.03 estimates in Hunt et al. Clinton and LeBoeuf 2000; see Table A3 (*) Elephant seal - Not present 0.00043 0.368 11.08 1993 See Table A2 (*) Dall’s porpoise 0.005986 0.005986 0.1 Kuzin et al. 2000 27.47 P. whitesided dolphin 0.003962 0.003962 0.14 Tanaka 1993 25.83 See Table A2 (*) N. right whale dolphin 0.003897 0.003897 0.16 Tanaka 1993 24.14 Common dolphin 0.001 - Not present 0.1 Hobbs and Jones 1993 24.98 Ogi et al. 1993 Albatross 0.00155 0.00004 0.05 81.59 (Midway Atoll) Shearwaters 0.001237 0.0004 0.1 100.1 Storm petrels 0.000157 0.000056 0.1 152.1 Kittiwakes 0.000115 Synthesis from Hunt 0.000052 Synthesis from Hunt 0.1 Average eastern Bering 123 Synthesis from Hunt et et al. -

Chinese Japanese Flying Squid (JFS) Fisheries Improvement Scoping Report
Chinese Japanese Flying Squid (JFS) Fisheries Improvement Scoping Report December 2018 Ocean Outcomes (O2) Contacts: Songlin Wang, China Program Director Qing Fang, Consultant Dr. Jocelyn Drugan, Analytics Team Director and Fishery Scientist Rich Lincoln, Founder and Senior Advisor www.oceanoutcomes.org 1 Chinese JFS Fishery Improvement Scoping Report: October 2018 TABLE OF CONTENTS 1. BACKGROUND .............................................................................................................................................. 4 1.1 OVERVIEW OF FISHERY PRE-ASSESSMENT .......................................................................................................................... 4 1.2 OVERVIEW OF FIP SCOPING .................................................................................................................................................... 5 2. STOCK AND FISHERY DESCRIPTION ...................................................................................................... 6 2.1 SPECIES AND STOCK .................................................................................................................................................................. 6 2.2 FISHERY OVERVIEW .................................................................................................................................................................. 9 2.2.1 Location ............................................................................................................................................................................ -

FAO Fisheries & Aquaculture
Food and Agriculture Organization of the United Nations Fisheries and for a world without hunger Aquaculture Department Species Fact Sheets Engraulis japonicus (Temminck & Schlegel, 1846) Black and white drawing: (click for more) Synonyms Engraulis japonicus Fowler, 1941d:694, (or incorrectly japonica ; Engraulis is masculine), (Japan, Korea, large synonymy). Atherina Japonica Houttuyn, 1782:340 (= nomen dubium - see Note below). Stolephorus celebicus Hardenberg, 1933a:262 (Manado, Sulawesi). FAO Names En - Japanese anchovy, Fr - Anchois japonais, Sp - Anchoíta japonesa. 3Alpha Code: JAN Taxonomic Code: 1210600202 Scientific Name with Original Description Engraulis japonicus Temminck & Schlegel, 1846, Fauna Japonica, Poiss., pt.13:239, pl. 108, fig.3 (southwest Japan). Diagnostic Features Differs very little from the European anchovy (see Engraulis encrasicolus ) and can be identified from that description. Of other anchovies found in the southern part of its distribution, only species of Encrasicholina and Stolephorus are similar in appearance (slender, rather round-bodied), but all have small spine like scutes before the pelvic fins (usually 2 to 7 scutes, but occasionally 1 or very rarely none in S. commersonii ). Species of Thryssa have compressed bodies and a keel of scutes along the belly. Geographical Distribution FAO Fisheries and Aquaculture Department Launch the Aquatic Species Distribution map viewer Western North and central Pacific (southern Sakhalin Island, Sea of Japan and Pacific coasts of Japan, and south to almost Canton/Taiwan -

A NOTE on the BIOLOGY and FISHERY of the JAPANESE ANCHOVY ENGRAULIS JAPONICA (HOUTTUYN) Slgeltl HAYASI Tokai Regional Fisheries Research Laboratory Tokyo, Japan
A NOTE ON THE BIOLOGY AND FISHERY OF THE JAPANESE ANCHOVY ENGRAULIS JAPONICA (HOUTTUYN) SlGElTl HAYASI Tokai Regional Fisheries Research laboratory Tokyo, Japan INTRODUCTION progress of the investigations. In addition, several individual biologists have presented comprehensive One of the most prolific groups of fishes in the papers on the Japanese anchovy, mainly in the pros- Japanese fisheries has been iwasi which comprises perous fishing areas. Hayasi (1961) , Asami (1962) three clupeoid species including the sardine, Sardi- and Takao (1964) discussed the investigations and nops meZanosticta (Temminck & Schlegel) , round her- management of the species and fisheries on the Pacific ring, Etrumeus micropus (Temminck & Schlegel) , and coasts of Honsyu, Sikoku and Kyusyu, and in the Set0 anchovy. These three species have been caught by Inland Sea, respectively. Kubo (1961) has given a various types of fishing methods in almost all of the review of works on the species under discussion. coastal waters around Japan, not only at stages after In spite of a number of investigations, however, it metamorphosis but also at postlarval stages under a is still difficult to assess and forecast the anchovy commercial name of sirasu, in combination with a few related fishes (Hayasi 1961). The total catch of iwasi population. Such deficit is noticed not only in the reached 1,130,000 metric tons, or 42 percent of the anchovy study but in the fishery biology of various total fish landings in Japan during 1929 through species (Ex. Com., Conf. Invest. Neritic-Pelagic 1938 (Nakai 1962b). Even though the catch of iwasi, Fisher. Japan, 1961-63, Sato 1961,1964).