Female, 28 and 22 Years)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Boards' Fodder
boards’ fodder Cosmeceuticals Contributed by Elisabeth Hurliman, MD, PhD; Jennifer Hayes, MD; Hilary Reich MD; and Sarah Schram, MD. INGREDIENT FUNCTION MECHANISM ASSOCIATIONS/SIDE EFFECTS Vitamin A/ Antioxidant (reduces free Affects gene transcription Comedolysis epidermal thickening, dermal Derivatives (retinal, radicals, lowers concentration differentiation and growth of regeneration, pigment lightening retinol, retinoic of matrix metalloproteinases cells in the skin acid, provitamin reduces collagen degradation) Side effects: Irritation, erythema, desquamation A, asthaxanthin, Normalizes follicular Elisabeth Hurliman, lutein) epithelial differentiation and keratinization MD, PhD, is a PGY-4 dermatology resident Vitamin C (L Secondary endogenous Ascorbic acid: necessary L-ascorbic acid + alpha-tocopherol (vitamin E)= ascorbic acid, antioxidant in skin cofactor for prolylhydroxylase UVA and UVB protection at University of tetrahexyldecyl and lysyl hydroxylase Minnesota department ascorbate) Lightens pigment Zinc, resveratrol, L-ergothioneine and tyrosine add of dermatology. (affects melanogenesis) L-ascorbic acid: scavenges to vitamin C bioavailability free oxygen radicals, Protects Vitamin E from oxidation stimulates collagen synthesis Improves skin texture and hydration May interrupt melanogenesis by interacting with copper ions Vitamin E/ Primary endogenous antioxidant Prevents lipid peroxidation; Alpha tocopherol is the most physiologically Tocopherols, in skin scavenges free oxygen active isomer Jennifer Hayes, MD, Tocotrienols -

The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (Cad) by Phosphorylation and Protein-Protein Interactions
THE REGULATION OF CARBAMOYL PHOSPHATE SYNTHETASE-ASPARTATE TRANSCARBAMOYLASE-DIHYDROOROTASE (CAD) BY PHOSPHORYLATION AND PROTEIN-PROTEIN INTERACTIONS Eric M. Wauson A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2007 Approved by: Lee M. Graves, Ph.D. T. Kendall Harden, Ph.D. Gary L. Johnson, Ph.D. Aziz Sancar M.D., Ph.D. Beverly S. Mitchell, M.D. 2007 Eric M. Wauson ALL RIGHTS RESERVED ii ABSTRACT Eric M. Wauson: The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (CAD) by Phosphorylation and Protein-Protein Interactions (Under the direction of Lee M. Graves, Ph.D.) Pyrimidines have many important roles in cellular physiology, as they are used in the formation of DNA, RNA, phospholipids, and pyrimidine sugars. The first rate- limiting step in the de novo pyrimidine synthesis pathway is catalyzed by the carbamoyl phosphate synthetase II (CPSase II) part of the multienzymatic complex Carbamoyl phosphate synthetase, Aspartate transcarbamoylase, Dihydroorotase (CAD). CAD gene induction is highly correlated to cell proliferation. Additionally, CAD is allosterically inhibited or activated by uridine triphosphate (UTP) or phosphoribosyl pyrophosphate (PRPP), respectively. The phosphorylation of CAD by PKA and ERK has been reported to modulate the response of CAD to allosteric modulators. While there has been much speculation on the identity of CAD phosphorylation sites, no definitive identification of in vivo CAD phosphorylation sites has been performed. Therefore, we sought to determine the specific CAD residues phosphorylated by ERK and PKA in intact cells. -

Proteomic Analysis of the Rad18 Interaction Network in DT40 – a Chicken B Cell Line
Proteomic analysis of the Rad18 interaction network in DT40 – a chicken B cell line Thesis submitted for the degree of Doctor of Natural Sciences at the Faculty of Biology, Ludwig-Maximilians-University Munich 15th January, 2009 Submitted by Sushmita Gowri Sreekumar Chennai, India Completed at the Helmholtz Zentrum München German Research Center for Environmental Health Institute of Clinical Molecular Biology and Tumor Genetics, Munich Examiners: PD Dr. Berit Jungnickel Prof. Heinrich Leonhardt Prof. Friederike Eckardt-Schupp Prof. Harry MacWilliams Date of Examination: 16th June 2009 To my Parents, Sister, Brother & Rajesh Table of Contents 1. SUMMARY ........................................................................................................................ 1 2. INTRODUCTION ............................................................................................................. 2 2.1. MECHANISMS OF DNA REPAIR ......................................................................................... 3 2.2. ADAPTIVE GENETIC ALTERATIONS – AN ADVANTAGE ....................................................... 5 2.3. THE PRIMARY IG DIVERSIFICATION DURING EARLY B CELL DEVELOPMENT ...................... 6 2.4. THE SECONDARY IG DIVERSIFICATION PROCESSES IN THE GERMINAL CENTER .................. 7 2.4.1. Processing of AID induced DNA lesions during adaptive immunity .................. 9 2.5. TARGETING OF SOMATIC HYPERMUTATION TO THE IG LOCI ............................................ 10 2.6. ROLE OF THE RAD6 PATHWAY IN IG DIVERSIFICATION -

Ligands of Therapeutic Utility for the Liver X Receptors
molecules Review Ligands of Therapeutic Utility for the Liver X Receptors Rajesh Komati, Dominick Spadoni, Shilong Zheng, Jayalakshmi Sridhar, Kevin E. Riley and Guangdi Wang * Department of Chemistry and RCMI Cancer Research Center, Xavier University of Louisiana, New Orleans, LA 70125, USA; [email protected] (R.K.); [email protected] (D.S.); [email protected] (S.Z.); [email protected] (J.S.); [email protected] (K.E.R.) * Correspondence: [email protected] Academic Editor: Derek J. McPhee Received: 31 October 2016; Accepted: 30 December 2016; Published: 5 January 2017 Abstract: Liver X receptors (LXRs) have been increasingly recognized as a potential therapeutic target to treat pathological conditions ranging from vascular and metabolic diseases, neurological degeneration, to cancers that are driven by lipid metabolism. Amidst intensifying efforts to discover ligands that act through LXRs to achieve the sought-after pharmacological outcomes, several lead compounds are already being tested in clinical trials for a variety of disease interventions. While more potent and selective LXR ligands continue to emerge from screening of small molecule libraries, rational design, and empirical medicinal chemistry approaches, challenges remain in minimizing undesirable effects of LXR activation on lipid metabolism. This review provides a summary of known endogenous, naturally occurring, and synthetic ligands. The review also offers considerations from a molecular modeling perspective with which to design more specific LXRβ ligands based on the interaction energies of ligands and the important amino acid residues in the LXRβ ligand binding domain. Keywords: liver X receptors; LXRα; LXRβ specific ligands; atherosclerosis; diabetes; Alzheimer’s disease; cancer; lipid metabolism; molecular modeling; interaction energy 1. -

Contig Protein Description Symbol Anterior Posterior Ratio
Table S2. List of proteins detected in anterior and posterior intestine pooled samples. Data on protein expression are mean ± SEM of 4 pools fed the experimental diets. The number of the contig in the Sea Bream Database (http://nutrigroup-iats.org/seabreamdb) is indicated. Contig Protein Description Symbol Anterior Posterior Ratio Ant/Pos C2_6629 1,4-alpha-glucan-branching enzyme GBE1 0.88±0.1 0.91±0.03 0.98 C2_4764 116 kDa U5 small nuclear ribonucleoprotein component EFTUD2 0.74±0.09 0.71±0.05 1.03 C2_299 14-3-3 protein beta/alpha-1 YWHAB 1.45±0.23 2.18±0.09 0.67 C2_268 14-3-3 protein epsilon YWHAE 1.28±0.2 2.01±0.13 0.63 C2_2474 14-3-3 protein gamma-1 YWHAG 1.8±0.41 2.72±0.09 0.66 C2_1017 14-3-3 protein zeta YWHAZ 1.33±0.14 4.41±0.38 0.30 C2_34474 14-3-3-like protein 2 YWHAQ 1.3±0.11 1.85±0.13 0.70 C2_4902 17-beta-hydroxysteroid dehydrogenase 14 HSD17B14 0.93±0.05 2.33±0.09 0.40 C2_3100 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 ABHD5 0.85±0.07 0.78±0.13 1.10 C2_15440 1-phosphatidylinositol phosphodiesterase PLCD1 0.65±0.12 0.4±0.06 1.65 C2_12986 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 PLCD1 0.76±0.08 1.15±0.16 0.66 C2_4412 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 PLCG2 1.13±0.08 2.08±0.27 0.54 C2_3170 2,4-dienoyl-CoA reductase, mitochondrial DECR1 1.16±0.1 0.83±0.03 1.39 C2_1520 26S protease regulatory subunit 10B PSMC6 1.37±0.21 1.43±0.04 0.96 C2_4264 26S protease regulatory subunit 4 PSMC1 1.2±0.2 1.78±0.08 0.68 C2_1666 26S protease regulatory subunit 6A PSMC3 1.44±0.24 1.61±0.08 -

Generated by SRI International Pathway Tools Version 25.0, Authors S
An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000238675-HmpCyc: Bacillus smithii 7_3_47FAA Cellular Overview Connections between pathways are omitted for legibility. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

Diplomarbeit
DIPLOMARBEIT Titel der Diplomarbeit „The influence of free fatty acids on the development of liver inflammation“ Verfasser Mario Kuttke, B.Sc. angestrebter akademischer Grad Magister der Naturwissenschaften (Mag.rer.nat.) Wien, 2012 Studienkennzahl lt. Studienblatt: A 490 Studienrichtung lt. Studienblatt: Diplomstudium Molekulare Biologie Betreuerin / Betreuer: A.o.Univ.-Prof.Dipl.-Ing.Dr. Marcela Hermann Danksagung Zuerst möchte ich mich bei a.o.Univ.-Prof. Dipl.-Ing. Dr. Marcela Hermann für die Betreuung meiner Diplomarbeit bedanken. Besonderer Dank gilt a.o.Univ-Prof. Dr. Bettina Grasl-Kraupp für die fachliche Unterstützung und Betreuung während der praktischen Durchführung der Arbeit. Weiters bedanke ich mich bei Sandra Sagmeister, Therese Böhm, Nora Bintner, Waltraud Schrottmaier, Melanie Pichlbauer, Marzieh Nejabat, Teresa Riegler, Bettina Wingelhofer und Christiane Maier für die ausgezeichnete Zusammenarbeit im Labor und die Unterstützung in allen Lebenslagen. Birgit Mir-Karner, Helga Koudelka und Krystyna Bukowska danke ich für ihre Hilfsbereitschaft und für die kollegiale Zusammenarbeit. Mein größter Dank gilt meinen Eltern, Ursula und Heinz, und meiner Großmutter, Theresia, die mir mein Studium ermöglicht und mich immer unterstützt haben, sowie meinem Bruder, Alex, der in allen Lebenslagen für mich da ist. Table of Contents TABLE OF CONTENTS INTRODUCTION ............................................................................................................................................. 4 HEPATOCELLULAR CARCINOMA (HCC) -

The Anti-Inflammatory Role of Nuclear Receptors in Dendritic Cells
The Anti-Inflammatory Role of Nuclear Receptors in Dendritic Cells A thesis submitted for the degree of Ph.D. By Mary Canavan B.Sc. (Hons), March 2012. Based on research carried out at School of Biotechnology, Dublin City University, Dublin 9, Ireland. Under the supervision of Dr. Christine Loscher. Declaration I hereby certify that this material, which I now submit for assessment on the programme of study leading to the award of Doctor of Philosophy is entirely my own work, that I have exercised reasonable care to ensure that the work is original, and does not to the best of my knowledge breach any law of copyright, and has not been taken from the work of others and to the extent that such work has been cited and acknowledged within the text of my work. Signed: ____________________ ID No.:__54351789__ Date: ______________ ACKNOWLEDGEMENTS There are so many people that I would like to thank and definitely not enough space to say exactly how grateful I am to you all. I have been lucky enough to work with an amazing group of people over the past few years. Firstly I would like to thank Christine for all your help, support, enthusiasm and patience – and for telling me not to do anymore of those p50 blots! I have thoroughly enjoyed working with you and learning from you over the last few years. To everyone in the Lab – you are the reason why I have such great memories when I look back at my time in DCU. Whenever I think of failed experiments, tough days and tears, there is always a great memory of you guys that goes along with it. -
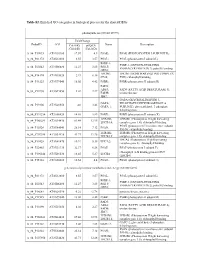
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4 -

Liver X Receptor &Beta
Cell Death and Differentiation (2014) 21, 1914–1924 & 2014 Macmillan Publishers Limited All rights reserved 1350-9047/14 www.nature.com/cdd Liver X receptor b activation induces pyroptosis of human and murine colon cancer cells V Derange`re1,2,3, A Chevriaux1,2, F Courtaut1,3, M Bruchard1,3, H Berger1,3, F Chalmin1,3, SZ Causse1, E Limagne1,3,FVe´gran1,3, S Ladoire1,2,3, B Simon4, W Boireau4, A Hichami1,3, L Apetoh1,2,3, G Mignot1, F Ghiringhelli1,2,3,5 and C Re´be´*,1,2,5 Liver X receptors (LXRs) have been proposed to have some anticancer properties, through molecular mechanisms that remain elusive. Here we report for the first time that LXR ligands induce caspase-1-dependent cell death of colon cancer cells. Caspase- 1 activation requires Nod-like-receptor pyrin domain containing 3 (NLRP3) inflammasome and ATP-mediated P2 Â 7 receptor activation. Surprisingly, LXRb is mainly located in the cytoplasm and has a non-genomic role by interacting with pannexin 1 leading to ATP secretion. Finally, LXR ligands have an antitumoral effect in a mouse colon cancer model, dependent on the presence of LXRb, pannexin 1, NLRP3 and caspase-1 within the tumor cells. Our results demonstrate that LXRb, through pannexin 1 interaction, can specifically induce caspase-1-dependent colon cancer cell death by pyroptosis. Cell Death and Differentiation (2014) 21, 1914–1924; doi:10.1038/cdd.2014.117; published online 15 August 2014 Liver X receptor a (LXRa) and b belong to the nuclear receptor However, a common feature of these reports is that all family. -

Fatty Acid Biosynthesis
BI/CH 422/622 ANABOLISM OUTLINE: Photosynthesis Carbon Assimilation – Calvin Cycle Carbohydrate Biosynthesis in Animals Gluconeogenesis Glycogen Synthesis Pentose-Phosphate Pathway Regulation of Carbohydrate Metabolism Anaplerotic reactions Biosynthesis of Fatty Acids and Lipids Fatty Acids contrasts Diversification of fatty acids location & transport Eicosanoids Synthesis Prostaglandins and Thromboxane acetyl-CoA carboxylase Triacylglycerides fatty acid synthase ACP priming Membrane lipids 4 steps Glycerophospholipids Control of fatty acid metabolism Sphingolipids Isoprene lipids: Cholesterol ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1 ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1. Biosynthesis of fatty acids 2. Regulation of fatty acid degradation and synthesis 3. Assembly of fatty acids into triacylglycerol and phospholipids 4. Metabolism of isoprenes a. Ketone bodies and Isoprene biosynthesis b. Isoprene polymerization i. Cholesterol ii. Steroids & other molecules iii. Regulation iv. Role of cholesterol in human disease ANABOLISM II: Biosynthesis of Fatty Acids & Lipids Lipid Fat Biosynthesis Catabolism Fatty Acid Fatty Acid Degradation Synthesis Ketone body Isoprene Utilization Biosynthesis 2 Catabolism Fatty Acid Biosynthesis Anabolism • Contrast with Sugars – Lipids have have hydro-carbons not carbo-hydrates – more reduced=more energy – Long-term storage vs short-term storage – Lipids are essential for structure in ALL organisms: membrane phospholipids • Catabolism of fatty acids –produces acetyl-CoA –produces reducing