Rdna) Organisation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
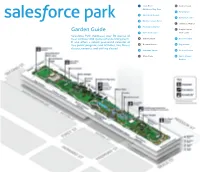
Salesforce Park Garden Guide
Start Here! D Central Lawn Children’s Play Area Garden Guide6 Palm Garden 1 Australian Garden Start Here! D Central Lawn Salesforce Park showcases7 California over Garden 50 species of Children’s Play Area 2 Mediterraneantrees and Basin over 230 species of understory plants. 6 Palm Garden -ã ¼ÜÊ ÊăØÜ ØÊèÜãE úØƀØÊèÃJapanese Maples ¼ÃØ Ê¢ 1 Australian Garden 3 Prehistoric¢ØÕ輫ÕØÊ£ØÂÜÃã«ó«ã«Üŧ¼«¹ĆãÃÜÜ Garden 7 California Garden ¼ÜÜÜŧÊÃØãÜŧÃØ¢ã«Ã£¼ÜÜÜũF Amphitheater Garden Guide 2 Mediterranean Basin 4 Wetland Garden Main Lawn E Japanese Maples Salesforce Park showcases over 50 species of 3 Prehistoric Garden trees and over 230 species of understory plants. A Oak Meadow 8 Desert Garden F Amphitheater It also offers a robust year-round calendar of 4 Wetland Garden Main Lawn free public programs and activities, like fitness B Bamboo Grove 9 Fog Garden Desert Garden classes, concerts, and crafting classes! A Oak Meadow 8 5 Redwood Forest 10 Chilean Garden B Bamboo Grove 9 Fog Garden C Main Plaza 11 South African 10 Chilean Garden Garden 5 Redwood Forest C Main Plaza 11 South African Garden 1 Children’s Australian Play Area Garden ABOUT THE GARDENS The botanist aboard the Endeavor, Sir Joseph Banks, is credited with introducing many plants from Australia to the western world, and many This 5.4 acre park has a layered soil system that plants today bear his name. balances seismic shifting, collects and filters storm- water, and irrigates the gardens. Additionally, the soil Native to eastern Australia, Grass Trees may grow build-up and dense planting help offset the urban only 3 feet in 100 years, and mature plants can be heat island effect by lowering the air temperature. -

Bush Foods and Fibres
Australian Plants Society NORTH SHORE GROUP Ku-ring-gai Wildflower Garden Bush foods and fibres • Plant-based bush foods, medicines and poisons can come from nectar, flowers, fruit, leaves, bark, stems, sap and roots. • Plants provide fibres and materials for making many items including clothes, cords, musical instruments, shelters, tools, toys and weapons. • A fruit is the seed-bearing structure of a plant. • Do not eat fruits that you do not know to be safe to eat. Allergic reactions or other adverse reactions could occur. • We acknowledge the Traditional Custodians of this land and pay our respects to the Elders both past, present and future for they hold the memories, traditions, culture and hope of their people. Plants as food: many native plants must be processed before they are safe to eat. Flowers, nectar, pollen, Sugars, vitamins, honey, lerps (psyllid tents) minerals, starches, manna (e.g. Ribbon Gum proteins & other nutrients Eucalyptus viminalis exudate), gum (e.g. Acacia lerp manna decurrens) Fruit & seeds Staple foods Carbohydrates (sugars, starches, fibre), proteins, fats, vitamins Leaves, stalks, roots, apical Staple foods Carbohydrates, protein, buds minerals Plants such as daisies, lilies, orchids and vines Tubers, rhyzomes were a source of starchy tubers known as Carbohydrate, fibre, yams. The yam daisy Microseris lanceolata protein, vitamins, (Asteraceae) was widespread in inland NSW minerals and other states. The native yam Dioscorea transversa grows north from Stanwell Tops into Qld and Northern Territory and can be eaten raw or roasted as can those of Trachymene incisa. 1 Plant Description of food Other notes Acacia Wattle seed is a rich source of iron, Saponins and tannins and other essential elements. -

Street Tree Master Plan Report © Sunshine Coast Regional Council 2009-Current
Sunshine Coast Street Tree Master Plan 2018 Part A: Street Tree Master Plan Report © Sunshine Coast Regional Council 2009-current. Sunshine Coast Council™ is a registered trademark of Sunshine Coast Regional Council. www.sunshinecoast.qld.gov.au [email protected] T 07 5475 7272 F 07 5475 7277 Locked Bag 72 Sunshine Coast Mail Centre Qld 4560 Acknowledgements Council wishes to thank all contributors and stakeholders involved in the development of this document. Disclaimer Information contained in this document is based on available information at the time of writing. All figures and diagrams are indicative only and should be referred to as such. While the Sunshine Coast Regional Council has exercised reasonable care in preparing this document it does not warrant or represent that it is accurate or complete. Council or its officers accept no responsibility for any loss occasioned to any person acting or refraining from acting in reliance upon any material contained in this document. Foreword Here on our healthy, smart, creative Sunshine Coast we are blessed with a wonderful environment. It is central to our way of life and a major reason why our 320,000 residents choose to live here – and why we are joined by millions of visitors each year. Although our region is experiencing significant population growth, we are dedicated to not only keeping but enhancing the outstanding characteristics that make this such a special place in the world. Our trees are the lungs of the Sunshine Coast and I am delighted that council has endorsed this master plan to increase the number of street trees across our region to balance our built environment. -

Complexity and Variation in the Effects of Low-Severity Fires on Forest Biota
University of Montana ScholarWorks at University of Montana Graduate Student Theses, Dissertations, & Professional Papers Graduate School 2003 Complexity and variation in the effects of low-severity fires on forest biota Karen Christine Short The University of Montana Follow this and additional works at: https://scholarworks.umt.edu/etd Let us know how access to this document benefits ou.y Recommended Citation Short, Karen Christine, "Complexity and variation in the effects of low-severity fires on forest biota" (2003). Graduate Student Theses, Dissertations, & Professional Papers. 9463. https://scholarworks.umt.edu/etd/9463 This Dissertation is brought to you for free and open access by the Graduate School at ScholarWorks at University of Montana. It has been accepted for inclusion in Graduate Student Theses, Dissertations, & Professional Papers by an authorized administrator of ScholarWorks at University of Montana. For more information, please contact [email protected]. Maureen and Mike MANSFIELD LIBRARY The University of Montana Permission is granted by the author to reproduce this material in its entirety, provided that this material is used for scholarly purposes and is properly cited in published works and reports. **Please check "Yes" or "No" and provide signature** ,/ Yes, I grant permission U7 ______ No, I do not grant permission Author's Signature: Date: <£/£-/< Any copying for commercial purposes or financial gain may be undertaken only with the author's explicit consent. 8/98 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. COMPLEXITY AND VARIATION IN THE EFFECTS OF LOW-SEVERITY FIRES ON FOREST BIOTA by Karen C. -

Yandina Street Tree Strategy
Yandina Street tree strategy Description of area and land use Canopy cover Street tree planting strategies The local plan area of Yandina occurs in the north of the Sunshine Coast Canopy cover over all lands is below-average for the region (31%) with Street trees enhance the historical look and feel of the township and Council region and totals 396 hectares in land area. The plan area contains the Foliage and Shade Cover plan for Yandina showing that open rural reinforce existing planting themes. the Yandina township, rural residential streets, farmlands, and industrial lands account for numerous areas of low or no tree cover. Vegetation cover and commercial precincts. Originally known as 'Native Dog Flat' the oldest reported for road reserve areas is also below average (27%). Analysis of Street tree planting focuses on shading pedestrian networks, building surveyed town in the Maroochy Shire was named Yandina in 1871. street tree occupancy within the town suggests that canopy cover can be canopy and establishing feature trees in key locations; and improving the readily increased through a solid program of proactive street tree planting. Yandina's landscape character beautifully blends the cultural heritage general amenity of town approaches. values of the small country town with the natural character of the area. Major opportunities and constraints The town's strong character tree palette bleeds out into surrounding Yellow flame trees frame the distinct facade of the village shop fronts while streets and links the sports precinct and other community facilities back clumps of eucalypts grow in areas immediately surrounding the township Numerous opportunities to build on the existing street tree canopy of to the town centre with feature and shade tree plantings. -

Brisbane Native Plants by Suburb
INDEX - BRISBANE SUBURBS SPECIES LIST Acacia Ridge. ...........15 Chelmer ...................14 Hamilton. .................10 Mayne. .................25 Pullenvale............... 22 Toowong ....................46 Albion .......................25 Chermside West .11 Hawthorne................. 7 McDowall. ..............6 Torwood .....................47 Alderley ....................45 Clayfield ..................14 Heathwood.... 34. Meeandah.............. 2 Queensport ............32 Trinder Park ...............32 Algester.................... 15 Coopers Plains........32 Hemmant. .................32 Merthyr .................7 Annerley ...................32 Coorparoo ................3 Hendra. .................10 Middle Park .........19 Rainworth. ..............47 Underwood. ................41 Anstead ....................17 Corinda. ..................14 Herston ....................5 Milton ...................46 Ransome. ................32 Upper Brookfield .......23 Archerfield ...............32 Highgate Hill. ........43 Mitchelton ...........45 Red Hill.................... 43 Upper Mt gravatt. .......15 Ascot. .......................36 Darra .......................33 Hill End ..................45 Moggill. .................20 Richlands ................34 Ashgrove. ................26 Deagon ....................2 Holland Park........... 3 Moorooka. ............32 River Hills................ 19 Virginia ........................31 Aspley ......................31 Doboy ......................2 Morningside. .........3 Robertson ................42 Auchenflower -

Bark Medicines Used in Traditional Healthcare in Kwazulu-Natal, South Africa: an Inventory
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector South African Journal of Botany 2003, 69(3): 301–363 Copyright © NISC Pty Ltd Printed in South Africa — All rights reserved SOUTH AFRICAN JOURNAL OF BOTANY ISSN 0254–6299 Bark medicines used in traditional healthcare in KwaZulu-Natal, South Africa: An inventory OM Grace1, HDV Prendergast2, AK Jäger3 and J van Staden1* 1 Research Centre for Plant Growth and Development, School of Botany and Zoology, University of Natal Pietermaritzburg, Private Bag X01, Scottsville 3209, South Africa 2 Centre for Economic Botany, Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3AE, United Kingdom 3 Department of Medicinal Chemistry, Royal Danish School of Pharmacy, 2 Universitetsparken, 2100 Copenhagen 0, Denmark * Corresponding author, e-mail: [email protected] Received 13 June 2002, accepted in revised form 14 March 2003 Bark is an important source of medicine in South Overlapping vernacular names recorded in the literature African traditional healthcare but is poorly documented. indicated that it may be unreliable in local plant identifi- From thorough surveys of the popular ethnobotanical cations. Most (43%) bark medicines were documented literature, and other less widely available sources, 174 for the treatment of internal ailments. Sixteen percent of species (spanning 108 genera and 50 families) used for species were classed in threatened conservation cate- their bark in KwaZulu-Natal, were inventoried. gories, but conservation and management data were Vernacular names, morphological and phytochemical limited or absent from a further 62%. There is a need for properties, usage and conservation data were captured research and specialist publications to address the in a database that aimed to synthesise published infor- gaps in existing knowledge of medicinal bark species mation of such species. -

Native Trees and Shrubs for Seaside Areas
NATIVE TREES & SHRUBS RECOMMENDED FOR SEASIDE AREAS NOOSA & DISTRICT LANDCARE GROUP STATION STREET, POMONA PH: 5485 2468 PLANTS SUITABLE FOR SEASIDE AREAS This list has been prepared as a guide to the selection of trees and shrubs suitable for planting on the coastal areas of Queensland. Most species are available from Noosa Landcare’s nurseries. Others listed are generally available from nurseries specializing in native plants. The problems of seaside plantings are mainly concerned with salt-laden winds which burn the foliage of tender plants. For such exposed sites the following plants will provide a resistant windbreak. SPECIES COMMON NAME (SHRUBS – Up to 5 metres) Acacia podalryiifolia Queensland silver wattle Acacia spectabilis Glory or Mudgee wattle Acacia suaveolens Sweet wattle Alectryon coriaceous Beach bird’s eye Baeckea frutescens Weeping baeckea Banksia ericiifolia Heath-leaved banksia Banksia oblingifolia Dwarf banksia Banksia spinulosa var. spinulosa Spiny-leaved honeysuckle Banksia spinulosa var. collina Hair-pin banksia Callistemon citrinus Crimson bottlebrush Callistemon pachyphyllus Wallum bottlebrush Callistemon rigidus Stiff bottlebrush Grevillea banksii Bank’s grevillea Hakea sericea White hakea Leptospermum brachyandrum Weeping tea-tree Leptospermum laevigatum Coast tea-tree Leptospermum petersonii Lemon-scented tea-tree Leptospermum polygalyfolium Wild may Melaleuca armillaris Bracelet honey myrtle Melaleuca alternifolia Paperbark Melaleauca bracteata River tea-tree Melaleuca nodosa Prickly-leaved paperbark Petalostigma -

Methods for Measuring Frost Tolerance of Conifers: a Systematic Map
Review Methods for Measuring Frost Tolerance of Conifers: A Systematic Map Anastasia-Ainhoa Atucha Zamkova *, Katherine A. Steele and Andrew R. Smith School of Natural Sciences, Bangor University, Bangor LL57 2UW, Gwynedd, UK; [email protected] (K.A.S.); [email protected] (A.R.S.) * Correspondence: [email protected] Abstract: Frost tolerance is the ability of plants to withstand freezing temperatures without unrecov- erable damage. Measuring frost tolerance involves various steps, each of which will vary depending on the objectives of the study. This systematic map takes an overall view of the literature that uses frost tolerance measuring techniques in gymnosperms, focusing mainly on conifers. Many different techniques have been used for testing, and there has been little change in methodology since 2000. The gold standard remains the field observation study, which, due to its cost, is frequently substituted by other techniques. Closed enclosure freezing tests (all non-field freezing tests) are done using various types of equipment for inducing artificial freezing. An examination of the literature indicates that several factors have to be controlled in order to measure frost tolerance in a manner similar to observation in a field study. Equipment that allows controlling the freezing rate, frost exposure time and thawing rate would obtain results closer to field studies. Other important factors in study design are the number of test temperatures used, the range of temperatures selected and the decrements between the temperatures, which should be selected based on expected frost tolerance of the tissue and species. Citation: Atucha Zamkova, A.-A.; Steele, K.A.; Smith, A.R. -

Ethiopia: the State of the World's Forest Genetic Resources
ETHIOPIA This country report is prepared as a contribution to the FAO publication, The Report on the State of the World’s Forest Genetic Resources. The content and the structure are in accordance with the recommendations and guidelines given by FAO in the document Guidelines for Preparation of Country Reports for the State of the World’s Forest Genetic Resources (2010). These guidelines set out recommendations for the objective, scope and structure of the country reports. Countries were requested to consider the current state of knowledge of forest genetic diversity, including: Between and within species diversity List of priority species; their roles and values and importance List of threatened/endangered species Threats, opportunities and challenges for the conservation, use and development of forest genetic resources These reports were submitted to FAO as official government documents. The report is presented on www. fao.org/documents as supportive and contextual information to be used in conjunction with other documentation on world forest genetic resources. The content and the views expressed in this report are the responsibility of the entity submitting the report to FAO. FAO may not be held responsible for the use which may be made of the information contained in this report. THE STATE OF FOREST GENETIC RESOURCES OF ETHIOPIA INSTITUTE OF BIODIVERSITY CONSERVATION (IBC) COUNTRY REPORT SUBMITTED TO FAO ON THE STATE OF FOREST GENETIC RESOURCES OF ETHIOPIA AUGUST 2012 ADDIS ABABA IBC © Institute of Biodiversity Conservation (IBC) -

SF Street Tree Species List 2019
Department of Public Works 2019 Recommended Street Tree Species List 1 Introduction The San Francisco Urban Forestry Council periodically reviews and updates this list of trees in collaboration with public and non-profit urban forestry stakeholders, including San Francisco Public Works, Bureau of Urban Forestry and Friends of the Urban Forest. The 2019 Street Tree List was approved by the Urban Forestry Council on October 22, 2019. This list is intended to be used for the public realm of streets and associated spaces and plazas that are generally under the jurisdiction of the Public Works. While the focus is on the streetscape, e.g., tree wells in the public sidewalks, the list makes accommodations for these other areas in the public realm, e.g., “Street Parks.” While this list recommends species that are known to do well in many locations in San Francisco, no tree is perfect for every potential tree planting location. This list should be used as a guideline for choosing which street tree to plant but should not be used without the help of an arborist or other tree professional. All street trees must be approved by Public Works before planting. Sections 1 and 2 of the list are focused on trees appropriate for sidewalk tree wells, and Section 3 is intended as a list of trees that have limited use cases and/or are being considered as street trees. Finally, new this year, Section 4, is intended to be a list of local native tree and arborescent shrub species that would be appropriate for those sites in the public realm that have more space than the sidewalk planting wells, for example, stairways, “Street Parks,” plazas, and sidewalk gardens, where more concrete has been extracted. -

Gymnosperms of Nilgiris District, Tamil Nadu
Research in Plant Biology, 4(6): 10-16, 2014 ISSN : 2231-5101 www.resplantbiol.com Regular Article Gymnosperms of Nilgiris District, Tamil Nadu *Jeevith S.1, Ramachandran V.S.1 and V. Ramsundar 2 1Taxonomy and Floristic Lab, Department of Botany, School of Life Sciences, Bharathiar University, Coimbatore - 641 046, Tamil Nadu, India, 2Government Botanical Garden, Udhagamandalam, Nilgiris - 643 001, Tamil Nadu, India * Corresponding author Email : [email protected] Gymnosperms are seed bearing vascular plants; an intermediate of Pteridophytes and Angiosperms occupying an important place in the plant kingdom. In the present study 43 species belonging to 20 genera comes under 10 families. The wild and exotic species are catalogue in Government botanical garden at Udhagamandalam for conservation strategies. Key words: Gymnosperms, naked seed, Nilgiris. A Gymnosperm is a seed plant that and they still successful in many parts of produces naked seeds that are not enclosed the world and occupy large of earth’s by a protective fruit. They have needle or surface (Dar and Dar, 2006). The scale like leaves and deep growing root importances of gymnosperms include system. Conifers are the largest group of mainly evergreen trees and shrubs, which gymnosperms on earth. The Gymnosperms are extremely captivating because of their are group co-ordinate with the graceful habit and attractive shapes Angiosperms within Phanerogams or (Poonam Tripati, Lalit M. Tewari, Ashish Spermatophytes. The Pteridospermales or Tewari, Sanjay Kumar, Pangtey, Y. P. S. and Cycadofilicales, fern like seed bearing plants Geeta Tewari, 2009). The Indian of Cordaitales dominated to the earth in subcontinent one of the 34 mega diversity Carboniferous and Permian periods.