Proteomic and Transcriptomic Study of Brain Microvessels in Neonatal And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Dataset for the Quantitative Proteomics Analysis of the Primary Hepatocellular Carcinoma with Single and Multiple Lesions
Data in Brief 5 (2015) 226–240 Contents lists available at ScienceDirect Data in Brief journal homepage: www.elsevier.com/locate/dib Data Article Dataset for the quantitative proteomics analysis of the primary hepatocellular carcinoma with single and multiple lesions Xiaohua Xing a,b, Yao Huang c, Sen Wang a,b, Minhui Chi a,b,c, Yongyi Zeng a,b,c, Lihong Chen a,b,c, Ling Li a,b,c, Jinhua Zeng a,b,c, Minjie Lin a,b, Xiao Han d, Jingfeng Liu a,b,c, Xiaolong Liu a,b,n a The Liver Center of Fujian Province, Fujian Medical University, Fuzhou 350025, People’s Republic of China b The United Innovation of Mengchao Hepatobiliary Technology Key Laboratory of Fujian Province, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou 350025, People’s Republic of China c Liver Disease Center, The First Affiliated Hospital of Fujian Medical University, Fuzhou 350007, People’s Republic of China d Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, People’s Republic of China article info abstract Article history: Hepatocellular Carcinoma (HCC) is one of the most common malig- Received 18 August 2015 nant tumor, which is causing the second leading cancer-related death Received in revised form worldwide. The tumor tissues and the adjacent noncancerous tissues 27 August 2015 obtained from HCC patients with single and multiple lesions were Accepted 28 August 2015 quantified using iTRAQ. A total of 5513 proteins (FDR of 1%) were Available online 8 September 2015 identified which correspond to roughly 27% of the total liver proteome. -

Hepatic Proteome Analysis Reveals Altered Mitochondrial Metabolism and Suppressed Acyl-Coa Synthetase-1 in Colon-26 Tumor-Induce
UCLA UCLA Previously Published Works Title Hepatic proteome analysis reveals altered mitochondrial metabolism and suppressed acyl- CoA synthetase-1 in colon-26 tumor-induced cachexia. Permalink https://escholarship.org/uc/item/5xb4k7mj Journal Physiological genomics, 52(5) ISSN 1094-8341 Authors Khamoui, Andy V Tokmina-Roszyk, Dorota Rossiter, Harry B et al. Publication Date 2020-05-01 DOI 10.1152/physiolgenomics.00124.2019 Peer reviewed eScholarship.org Powered by the California Digital Library University of California 1 Hepatic proteome analysis reveals altered mitochondrial metabolism and suppressed acyl-CoA 2 synthetase-1 in colon-26 tumor-induced cachexia 3 4 Andy V. Khamoui1,2*, Dorota Tokmina-Roszyk2,3, Harry B. Rossiter5,6, Gregg B. Fields2,3,4, Nishant P. 5 Visavadiya1 6 7 1Department of Exercise Science and Health Promotion, Florida Atlantic University, Boca Raton, FL, USA; 8 2Institute for Human Health & Disease Intervention, Florida Atlantic University, Boca Raton, FL, USA; 9 3Department of Chemistry & Biochemistry, Florida Atlantic University, Jupiter, FL, USA; 4Department of 10 Chemistry, The Scripps Research Institute, Jupiter, FL, USA; 5Division of Respiratory and Critical Care 11 Physiology and Medicine, Department of Medicine, The Lundquist Institute for Biomedical Innovation at 12 Harbor-UCLA Medical Center, Torrance, CA, USA; 6Faculty of Biological Sciences, University of Leeds, Leeds, 13 UK 14 15 Running Title: Hepatic proteome in cancer cachexia 16 17 *Correspondence 18 Andy V. Khamoui PhD 19 Department of Exercise Science and Health Promotion 20 Florida Atlantic University 21 777 Glades Rd, FH-11A, Rm 128-B 22 Boca Raton, FL 33431 23 Tel: (561) 297-4450 | Email: [email protected] 24 25 26 27 28 1 Downloaded from journals.physiology.org/journal/physiolgenomics by Harry Rossiter (047.144.146.194) on March 14, 2020. -
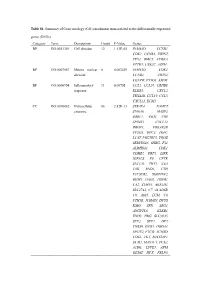
Table S1. Summary of Gene Ontology (GO) Enrichment Terms Related To
Table S1. Summary of Gene ontology (GO) enrichment terms related to the differentially expressed genes (DEGs) Category Term Description Count P-Value Genes BP GO:0051301 Cell division 12 1.13E-04 FAM83D, CCNB1, CDK1, CCNB2, ZWINT, TPX2, BIRC5, AURKA, PTTG1, UBE2C, ASPM BP GO:0007067 Mitotic nuclear 8 0.003429 FAM83D, CDK1, division CCNB2, ZWINT, CENPW, PTTG1, ASPM BP GO:0006954 Inflammatory 11 0.00705 CCL2, CCL20, CRHBP, response KLKB1, CXCL2, TBXA2R, CCL19, CCL5, CXCL12, ECM1 CC GO:0070062 Extracellular 86 2.82E-13 STEAP4, NAMPT, exosome S100A8, MASP2, SORL1, ANO1, CRP, SPINK1, CXCL12, BBOX1, PRKAR2B, TP53I3, GPC3, TKFC, LCAT, PGLYRP2, PROZ, SERPINA4, SHBG, F11, ALDH6A1, CDK1, CDHR2, FBP1, LIFR, IGFALS, F9, CFTR, SLC3A1, THY1, C8A, C8B, RND3, CTH, TACSTD2, SERPINF2, BHMT, HAO2, HSPB1, CA2, STMN1, AKR1D1, SLC27A2, C7, ACADSB, C9, ASS1, LUM, C6, PTH1R, JCHAIN, DPYS, KMO, SFN, ARG1, ANGPTL6, KLKB1, ENO3, HRG, SLC39A5, SPP2, SPP1, DPT, THBS4, GPD1, HOGA1, EPHX2, FTCD, ACMSD, CD5L, TKT, RACGAP1, ECM1, MAN1C1, PCK1, A1BG, LYVE1, AFM, GLYAT, HPX, FBLN5, PON1, CP, HPGD, IGFBP3, FABP5 CC GO:0072562 Blood 13 7.61E-09 C9, JCHAIN, CD5L, microparticle C8A, A1BG, AFM, BCHE, HPX, SERPINF2, APOA5, PON1, HRG, CP CC GO:0005615 Extracellular 39 2.25E-06 XDH, MBL2, CCL2, space S100A8, ADAMTS13, CRHBP, LUM, CRP, CXCL2, SORL1, CD109, SPINK1, CCL5, PTGIS, CCL20, BCHE, ANG, KLKB1, PROZ, SERPINA4, ENO3, NRG1, SPP1, THBS4, DPT, F11, IGF1, CCL19, F9, DBH, ECM1, BMPER, SERPINF2, TACSTD2, FBLN5, PON1, HSPB1, IGFBP3 CC GO:0005739 Mitochondrion 29 0.002339 ETNPPL, -

Supporting Information
Supporting Information Table S1. List of confirmed SLC transporters represented in Canine GeneChip. SLC family Members detected Members not detected SLC1: The high affinity glutamate and neutral amino acid SLC1A1 SLC1A2, SLC1A3, SLC1A6 transporter family SLC2: The facilitative GLUT transporter family SLC2A1, SLC2A8 SLC2A3, SLC2A9 SLC3: The heavy subunits of the heteromeric amino acid SLC3A1 transporters SLC4: The bicarbonate transporter family SLC4A11 SLC4A4, SLC4A8 SLC5: The sodium glucose cotransporter family SLC5A6 SLC5A3, SLC5A10, SLC5A12 SLC6: The sodium- and chloride- dependent SLC6A6, SLC6A12 SLCA18 neurotransmitter transporter family SLC7: The cationic amino acid transporter/glycoprotein- NR associated family SLC8: The Na+/Ca2+ exchanger family SLC8A1 SLC9: The Na+/H+ exchanger family SLC9A1, SLC9A6, SLC9A9 SLC10: The sodium bile salt cotransport family SLC10A2 SLC11: The proton coupled metal ion transporter family NR SLC12: The electroneutral cation-Cl cotransporter family SLC12A3, SLC12A6, SLC12A8 SLC13: The human Na+-sulfate/carboxylate cotransporter SLC13A2 family SLC14: The urea transporter family NR SLC15: The proton oligopeptide cotransporter family SLC15A2, SLC15A4 SLC15A1 SLC16: The monocarboxylate transporter family SLC16A13 SLC16A4 SLC17: The vesicular glutamate transporter family SLC17A3, SLC17A7 SLC18: The vesicular amine transporter family NR SLC19: The folate/thiamine transporter family NR SLC20: The type III Na+-phosphate cotransporter family NR SLC21/SLCO: The organic anion transporting family SLC21A3, SLC21A8, -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 7, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on September 29, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009. -

Metabolic Network-Based Stratification of Hepatocellular Carcinoma Reveals Three Distinct Tumor Subtypes
Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes Gholamreza Bidkhoria,b,1, Rui Benfeitasa,1, Martina Klevstigc,d, Cheng Zhanga, Jens Nielsene, Mathias Uhlena, Jan Borenc,d, and Adil Mardinoglua,b,e,2 aScience for Life Laboratory, KTH Royal Institute of Technology, SE-17121 Stockholm, Sweden; bCentre for Host-Microbiome Interactions, Dental Institute, King’s College London, SE1 9RT London, United Kingdom; cDepartment of Molecular and Clinical Medicine, University of Gothenburg, SE-41345 Gothenburg, Sweden; dThe Wallenberg Laboratory, Sahlgrenska University Hospital, SE-41345 Gothenburg, Sweden; and eDepartment of Biology and Biological Engineering, Chalmers University of Technology, SE-41296 Gothenburg, Sweden Edited by Sang Yup Lee, Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea, and approved November 1, 2018 (received for review April 27, 2018) Hepatocellular carcinoma (HCC) is one of the most frequent forms of of markers associated with recurrence and poor prognosis (13–15). liver cancer, and effective treatment methods are limited due to Moreover, genome-scale metabolic models (GEMs), collections tumor heterogeneity. There is a great need for comprehensive of biochemical reactions, and associated enzymes and transporters approaches to stratify HCC patients, gain biological insights into have been successfully used to characterize the metabolism of subtypes, and ultimately identify effective therapeutic targets. We HCC, as well as identify drug targets for HCC patients (11, 16–18). stratified HCC patients and characterized each subtype using tran- For instance, HCC tumors have been stratified based on the uti- scriptomics data, genome-scale metabolic networks and network lization of acetate (11). Analysis of HCC metabolism has also led topology/controllability analysis. -

Type of the Paper (Article
The Consumption of Cholesterol-Enriched Diets Conditions the Development of a Subtype of HCC with High Aggressiveness and Poor Prognosis Arturo Simoni-Nieves, Soraya Salas-Silva, Lisette Chávez-Rodríguez, Alejandro Escobedo-Calvario, Matthis Desoteux, Leticia Bucio, Verónica Souza, Roxana U. Miranda-Labra, Linda E. Muñoz-Espinosa, Cédric Coulouarn, María Concepción Gutiérrez-Ruiz, Jens U. Marquardt and Luis E. Gomez-Quiroz Figure S1. A) Gross inspection of mice fed with the different diets and treated or not with DEN. Representative image of at least eight animals, white arrows show adipose tissue. B) Tumor gross inspection of mice under different diets. Repre- sentative image of at least eight livers, yellow arrows show vascularity. Images are representative of at least eight mice. Cancers 2021, 13, 1721. https://doi.org/10.3390/cancers13071721 www.mdpi.com/journal/cancers Cancers 2021, 13, 1721 2 of 29 Figure S2. Expression of eight randomly selected genes in samples not included in the RNA-seq, as technical and biological validation. Each graph plots the individual data points, the superimpose horizontal line indicates the arithmetic mean and error bars showing ± SEM. ** p < 0.01 vs CW group; & p < 0.05 vs diet control. Cancers 2021, 13, 1721 3 of 29 Figure S3. Expression of the 4 genes differentially overexpressed related to high lipid overload with potential relevance in tumor promotion: Mthfd1, Slc41a3, Fabp5, Igdcc4. Each graph plots the individual data points, the superimpose hori- zontal line indicates the arithmetic mean and error bars showing ± SEM. ** p < 0.01 vs CW group. Table S1. Mice groups under different diets and treatments. -

RNA-Seq Reveals Conservation of Function Among the Yolk Sacs Of
RNA-seq reveals conservation of function among the PNAS PLUS yolk sacs of human, mouse, and chicken Tereza Cindrova-Daviesa, Eric Jauniauxb, Michael G. Elliota,c, Sungsam Gongd,e, Graham J. Burtona,1, and D. Stephen Charnock-Jonesa,d,e,1,2 aCentre for Trophoblast Research, Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 3EG, United Kingdom; bElizabeth Garret Anderson Institute for Women’s Health, Faculty of Population Health Sciences, University College London, London, WC1E 6BT, United Kingdom; cSt. John’s College, University of Cambridge, Cambridge, CB2 1TP, United Kingdom; dDepartment of Obstetrics and Gynaecology, University of Cambridge, Cambridge, CB2 0SW, United Kingdom; and eNational Institute for Health Research, Cambridge Comprehensive Biomedical Research Centre, Cambridge, CB2 0QQ, United Kingdom Edited by R. Michael Roberts, University of Missouri-Columbia, Columbia, MO, and approved May 5, 2017 (received for review February 14, 2017) The yolk sac is phylogenetically the oldest of the extraembryonic yolk sac plays a critical role during organogenesis (3–5, 8–10), membranes. The human embryo retains a yolk sac, which goes there are limited data to support this claim. Obtaining experi- through primary and secondary phases of development, but its mental data for the human is impossible for ethical reasons, and importance is controversial. Although it is known to synthesize thus we adopted an alternative strategy. Here, we report RNA proteins, its transport functions are widely considered vestigial. sequencing (RNA-seq) data derived from human and murine yolk Here, we report RNA-sequencing (RNA-seq) data for the human sacs and compare them with published data from the yolk sac of and murine yolk sacs and compare those data with data for the the chicken. -

ADX609 3301 Athena Newborndx Gene Panel Update 3-30-20.Indd
NewbornDx™ Advanced Sequencing Evaluation When time to diagnosis matters, the NewbornDx™ Advanced Sequencing Evaluation from Athena Diagnostics delivers rapid, 3- to 7-day results on a targeted 1,722-genes. A2ML1 ALAD ATM CAV1 CLDN19 CTNS DOCK7 ETFB FOXC2 GLUL HOXC13 JAK3 AAAS ALAS2 ATP1A2 CBL CLIC2 CTRC DOCK8 ETFDH FOXE1 GLYCTK HOXD13 JUP AARS2 ALDH18A1 ATP1A3 CBS CLMP CTSA DOK7 ETHE1 FOXE3 GM2A HPD KANK1 AASS ALDH1A2 ATP2B3 CC2D2A CLN3 CTSD DOLK EVC FOXF1 GMPPA HPGD K ANSL1 ABAT ALDH3A2 ATP5A1 CCDC103 CLN5 CTSK DPAGT1 EVC2 FOXG1 GMPPB HPRT1 KAT6B ABCA12 ALDH4A1 ATP5E CCDC114 CLN6 CUBN DPM1 EXOC4 FOXH1 GNA11 HPSE2 KCNA2 ABCA3 ALDH5A1 ATP6AP2 CCDC151 CLN8 CUL4B DPM2 EXOSC3 FOXI1 GNAI3 HRAS KCNB1 ABCA4 ALDH7A1 ATP6V0A2 CCDC22 CLP1 CUL7 DPM3 EXPH5 FOXL2 GNAO1 HSD17B10 KCND2 ABCB11 ALDOA ATP6V1B1 CCDC39 CLPB CXCR4 DPP6 EYA1 FOXP1 GNAS HSD17B4 KCNE1 ABCB4 ALDOB ATP7A CCDC40 CLPP CYB5R3 DPYD EZH2 FOXP2 GNE HSD3B2 KCNE2 ABCB6 ALG1 ATP8A2 CCDC65 CNNM2 CYC1 DPYS F10 FOXP3 GNMT HSD3B7 KCNH2 ABCB7 ALG11 ATP8B1 CCDC78 CNTN1 CYP11B1 DRC1 F11 FOXRED1 GNPAT HSPD1 KCNH5 ABCC2 ALG12 ATPAF2 CCDC8 CNTNAP1 CYP11B2 DSC2 F13A1 FRAS1 GNPTAB HSPG2 KCNJ10 ABCC8 ALG13 ATR CCDC88C CNTNAP2 CYP17A1 DSG1 F13B FREM1 GNPTG HUWE1 KCNJ11 ABCC9 ALG14 ATRX CCND2 COA5 CYP1B1 DSP F2 FREM2 GNS HYDIN KCNJ13 ABCD3 ALG2 AUH CCNO COG1 CYP24A1 DST F5 FRMD7 GORAB HYLS1 KCNJ2 ABCD4 ALG3 B3GALNT2 CCS COG4 CYP26C1 DSTYK F7 FTCD GP1BA IBA57 KCNJ5 ABHD5 ALG6 B3GAT3 CCT5 COG5 CYP27A1 DTNA F8 FTO GP1BB ICK KCNJ8 ACAD8 ALG8 B3GLCT CD151 COG6 CYP27B1 DUOX2 F9 FUCA1 GP6 ICOS KCNK3 ACAD9 ALG9 -

Transporters
University of Dundee The Concise Guide to PHARMACOLOGY 2015/16 Alexander, Stephen P. H.; Kelly, Eamonn; Marrion, Neil; Peters, John A.; Benson, Helen E.; Faccenda, Elena Published in: British Journal of Pharmacology DOI: 10.1111/bph.13355 Publication date: 2015 Licence: CC BY Document Version Publisher's PDF, also known as Version of record Link to publication in Discovery Research Portal Citation for published version (APA): Alexander, S. P. H., Kelly, E., Marrion, N., Peters, J. A., Benson, H. E., Faccenda, E., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A., & CGTP Collaborators (2015). The Concise Guide to PHARMACOLOGY 2015/16: Transporters. British Journal of Pharmacology, 172(24), 6110-6202. https://doi.org/10.1111/bph.13355 General rights Copyright and moral rights for the publications made accessible in Discovery Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from Discovery Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain. • You may freely distribute the URL identifying the publication in the public portal. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 06. Oct. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: Transporters.