Osbhlh073 Negatively Regulates Internode Elongation and Plant Height by Modulating GA Homeostasis in Rice
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evaluating Ethylene Sensitivity Using Mature Plant Screens and the Seedling Hypocotyl Response
Evaluating Ethylene Sensitivity Using Mature Plant Screens and the Seedling Hypocotyl Response THESIS Presented in Partial Fulfillment of the Requirements for the Degree Master of Science in the Graduate School of The Ohio State University By Nichole Frances Edelman, B. A. Graduate Program in Horticulture and Crop Science The Ohio State University 2013 Master's Examination Committee: Dr. Michelle L. Jones, Advisor Dr. Pablo Jourdan Dr. Claudio Pasian Copyrighted by Nichole Frances Edelman 2013 Abstract Ethylene (C2H4) is a gaseous hormone produced by plants in response to environmental stress and during growth and development. Sensitive seedlings exposed to ethylene gas will exhibit an exaggerated apical hook, thickened hypocotyl, and reduced hypocotyl elongation. Collectively these symptoms are known as the triple response, and have been used as a screen to identify ethylene mutants. Exposure to ethylene gas at the mature plant stage can induce flower, bud, or leaf abscission, flower senescence, leaf chlorosis (yellowing), or leaf epinasty (downward curvature). Sensitivity can vary between species and by developmental stage or tissue (flower vs leaf). Ethylene can damage plants during production, shipping, and retailing. The objectives of this research were 1) to determine if the seedling hypocotyl elongation screen could be used to predict the sensitivity of mature plants at the marketable stage; 2) to identify ethylene sensitivity differences (levels of sensitivity and symptoms) between accessions within Solanaceae; and 3) to identify ethylene sensitivity differences at different developmental stages (seedling, juvenile, and mature plants). Seedlings were germinated on filter paper saturated with 1- aminocyclopropane-1-carboxylic acid (ACC) and sensitivity was determined based on the hypocotyl lengths relative to the control (0 µM ACC). -
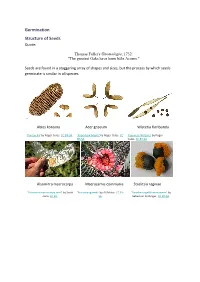
Germination Structure of Seeds Quote
Germination Structure of Seeds Quote: Thomas Fuller's Gnomologia, 1732: "The greatest Oaks have been little Acorns." Seeds are found in a staggering array of shapes and sizes, but the process by which seeds germinate is similar in all species. Abies koreana Acer griseum Wisteria floribunda 'Korean Fir' by Roger Culos. CC BY-SA. 'Paperbark Maple' by Roger Culos. CC 'Japanese Wisteria' by Roger BY-SA. Culos. CC BY-SA. Alsomitra macrocarpa Macrozamia communis Strelitzia reginae 'Alsomitra macrocarpa seed' by Scott 'BurrawangSeeds' by AYArktos. CC BY- 'Paradiesvogelblumensamen' by Zona. CC BY. SA. Sebastian Stabinger. CC BY-SA. Taraxicum officinale Stephanotis floribunda Phleum pratense 'Achane of Taraxacum sect. 'Stephanotis seed' by L. Marie"/Lenore 'Timoteegras vruchten Phleum Ruderalia' by Didier Edman, Sunnyvale, CA. CC BY. pratense' by Rasbak. CC BY-SA. Descouens. CC BY-SA. Dicotyledon seeds testa epicotyl plumule hypocotyl cotyledon radicle 'Aesculus hippocastanum seed section' by Boronian. CC BY. plumule epicotyl hypocotyl testa hilum radicle cotyledon micropyle endosperm Monocotyledon seeds endosperm epicotyl testa hypocotyl cotyledon radicle Parts of a seed Testa The seed coat. A protective layer which is tough and hard and it protects the seed from attack by insects, fungi and bacteria. Cotyledon Dicotyledons have 2 cotyledons Monocotyledons have 1 cotyledon A cotyledon is an embryonic leaf. It is the first leaf to appear when a seedling grows. They often contain reserves of food which the developing seedling can use to grow. Epicotyl The section of stem between the cotyledon(s) and the plumule. In a seedling it is the section of stem between the cotyledons and the first true leaves. -
4-H Growing Things Series Discovering the Science of Plants Welcome 4-H Leaders!
4-H Growing Things Series Discovering the Science of Plants Welcome 4-H Leaders! Welcome to the “Discovering the Science of Plants” project. There is lots of information, fun facts and hands on activities that cover the basic scientific Table of Contents principles of how plants grow and survive. This guide provides you with Introduction 1 project meeting plans (Skill Builders) that include, a skills list, background information, activity suggestions, and ways to know if your members have Project Summary 2 learned the skills identified. Skill Builder 1: 5 What is a plant? In this project, members will examine, by learning to do by doing, how plants grow and how they are used in everyday life. The Leader Guide is written with Skill Builder 2: 11 the expectation that the project leader(s) will have a working knowledge about Seeds for the the project topics and how they work. If not, you may need to do some Future pre-work / research on the activities, or recruit assistance for certain sections. Skill Builder 3: 18 Getting to the Be sure to try out activities, demonstrations or hands on work ahead of time to Root of it ensure you have an understanding of each Skill Builder - this also allows for any adjustments should an activity not work for you or if any equipment or supplies Skill Builder 4: 26 are unavailable. Leaves and Stems: the Power The 3D’s of Learning - Each Skill Builder has three sections of learning called Generators “Dream it!”, “Do it!” and “Dig it!”. Below is a description of each. -

Tulipa Kaufmanniana Regel.) Propagation Under Ex Situ Conditions ISSN 2447-536X |
THE APPLICATION OF DIFFERENT REPRODUCTION TECHNIQUES FOR RARE SPECIES WATERLILY TULIP 450 Ornamental (HorticultureTULIPA KAUFMANNIANA REGEL.) PROPAGATION UNDER EX SITU CONDITIONS ISSN 2447-536X | HTTPS://ORNAMENTALHORTICULTURE.EMNUVENS.COM.BR/RBHO SCIENTIFIC ARTICLE The application of different reproduction techniques for rare species waterlily tulip (Tulipa kaufmanniana Regel.) propagation under ex situ conditions Nabieva Alexandra Yurievna1*, Gerasimovich Lyudmila Vladimirovna2 Abstract T. kaufmanniana is a threatened and endemic species of the Tien Shan Mountains with a complex of valuable ornamental features. However, the commercial usage of the plant is limited due to the restricted availability of the bulbs in nature, difficulties in overcoming seed dormancy and low efficiency of the species reproduction. Results of the investigation of the reproductive biology of this species in the culture conditions allowed us to characterize T. kaufmanniana as viable with successful seedage. In the work reported here the effect of low temperature on the proper development of embryos is addressed, and an attempt is made to clarify the effect of cytokinins (TDZ, BAP) and auxin (NAA) on the regeneration capacity of isolated T. kaufmanniana embryos for the adventitious shoots and bulblets formation. Direct shoot organogenesis was induced by the combined action of chilling and BAP or TDZ treatment. Among the variants tested, TDZ promoted the formation of adventitious bulbs in 48 % of the chilled embryos, compared to non-chilled embryos (0.0%) and embryos exposed to the same concentration of BAP (39%). The best culture condition for T. kaufmanniana embryos growth and bulblets formation consisted of chilling for 10 wk in the dark at 4 °C, 4 wk at 20 °C under 16 h light photoperiod and 10 wk at 4 °C in the dark. -

Early Soybean Growth and Development
EARLY SOYBEAN GROWTH AND DEVELOPMENT A soybean seed has two distinct parts: the cotyledons and the embryo. The two cotyledons are the main food storage structure, which supply food during emergence and for the seven to ten days after emergence through the V1 growth stage. The embryo itself is comprised of three parts. 1. The radicle is the first root and it’s the first part of the embryo to penetrate the seed coat. Lateral roots form from the radicle, and tiny root hairs then develop on the lateral roots. Root hairs are the main absorbing surface of the entire root system. A soybean’s root system branches and rebranches within the first four to five weeks after planting, with the bulk of root mass persisting in the top six inches of soil. 2. The hypocotyl is the stem below the cotyledon. It begins to elongate after the radicle and forms an arch, which is pushed upward. As the arch breaks the soil surface, it pulls the cotyledons and epicotyl up. The uppermost cells of the hypocotyl stop growing as cells on the underside continue to straighten the arch. This process lifts the cotyledons into an upright position. 3. The epicotyl is comprised of the small leaves, stem and the apical growing point. The epicotyl is protected between the cotyledons until after emergence. The loss of a cotyledon is not lethal to the seedling, but damage to the epicotyl is. The hypocotyl arch is a large structure that is relative to seed size, utilizing considerable seed energy to push through the soil. -

The Effect of Light Intensity on the Hypocotyl Length of Arabidopsis Thaliana Christina Bell, Mandy Choi, Audrey Lam, and Kimber
The effect of light intensity on the hypocotyl length of Arabidopsis thaliana Christina Bell, Mandy Choi, Audrey Lam, and Kimberley Xiao Abstract The amount of light a plant receives during germination can greatly affect its growth and morphology. In this study, seeds of Arabidopsis thaliana were used to investigate the effects of light intensity on hypocotyl growth and length. Seeds of both the wild type and the mutant cer10 of Arabidopsis thaliana were grown in petri dishes for a period of eleven days, under three varying light intensities. These were marked no light, low light and high light, averaging at 3 lux, 141 lux, and 6969 lux, respectively. It was found that for both the mutant and the wild type, seeds grown in a no light environment had a significantly longer hypocotyl (embryonic stem) length than those grown at high light intensity. For the wild type, at the end of the 11th day, those grown in the high light treatment had a mean hypocotyl length of 0.7±0.05mm, those grown in the low light treatment had a mean hypocotyl length of 4.0±0.8mm, and those grown in the no light treatment had a mean hypocotyl length of 10.5±1.3mm. For the mutant cer10, the high light treatment had a mean hypocotyl length of 1.0±0.2mm, the low light treatment had a mean hypocotyl length of 4.0±0.8mm, and the no light treatment had a mean hypocotyl length of 11.8±0.6mm. The results agree with current literature on the subject; however we fail to reject our initial null hypothesis for both the mutant and wild type. -

Soybean Temperature-Dependent Hypocotyl Growth Anomaly: Physiological Aspects Robert Dean Keys Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1979 Soybean temperature-dependent hypocotyl growth anomaly: physiological aspects Robert Dean Keys Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Botany Commons Recommended Citation Keys, Robert Dean, "Soybean temperature-dependent hypocotyl growth anomaly: physiological aspects " (1979). Retrospective Theses and Dissertations. 6651. https://lib.dr.iastate.edu/rtd/6651 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This was produced from a copy of a document sent to us for microfîlming. While the most advanced technological means to photograph and reproduce this document have been used, the quality is heavily dependent upon the quality of the material submitted. The following explanation of techniques is provided to help you understand markings or notations which may appear on this reproduction. 1. The sign or "target" for pages apparently lacking from the document photographed is "Missing Page(s)". If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting through an image and duplicating adjacent pages to assure you of complete continuity. 2. When an image on the film is obliterated with a round black mark it is an indication that the film inspector noticed either blurred copy because of movement during exposure, or duplicate copy. -

Anthocyanin Accumulation in the Hypocotyl and Petal of Red Agati (Sesbania Grandiflora), an Ornamental Legume
Journal Journal of Applied Horticulture, 8(2): 143-146, July-December, 2006 Appl Anthocyanin accumulation in the hypocotyl and petal of Red Agati (Sesbania grandiflora), an ornamental legume Kitti Bodhipadma1, Sompoch Noichinda1, Sasikan Udomrati1, Goravis Nathalang2, Boonyuen Kijwijan3 and David W.M. Leung4 1Department of Agro-Industrial Technology, Faculty of Applied Science, King Mongkut’s Institute of Technology North Bangkok, Bangsue, Bangkok, 10800, Thailand; 2Department of Biology, Faculty of Science, Mahidol University, Bang- kok, 10400, Thailand; 3Department of Biology, Faculty of Science, Khon Kaen University, Khon Kaen, 40002, Thailand; 4School of Biological Sciences, University of Canterbury, Private Bag 4800, Christchurch 1, New Zealand, E-mail: david. [email protected] Abstract Seeds of Red Agati (Sesbania grandiflora), an ornamental leguminous tree, were germinated in vitro under both light and dark conditions for 7, 10, 15, 20 and 25 days. The localization of anthocyanin-containing cells and level of total anthocyanin content of hypocotyl from several developmental stages were determined. In the hypocotyl of light-grown seedlings, anthocyanin-containing cells were observed in epidermal and sub-epidermal layer and peripheral cortex while none was found in that of dark-grown seedlings. On day 7, the hypocotyl of light-grown seedlings had the highest anthocyanin content (290 μg/g FW). Moreover, Red Agati’s petal at various developmental stages was also examined for the total anthocyanin content. It was found that the petal of 3 cm length had the highest total anthocyanin level (455 μg/g FW). It is concluded that the hypocotyl of light-grown Red Agati seedlings is an attractive alternative source of anthocyanins to the petal as the seedlings can be raised and be made available throughout the year. -

Investigating Hormone Regulation and Sugar Storage During Tuber Development in Turnip Plants (Brassica Rapa)
Investigating Hormone Regulation and Sugar Storage during Tuber Development in Turnip Plants (Brassica rapa) MSc Thesis Report (PBR-80436) Temesgen Menamo (Reg. No. 860717-557-050) Supervisors: Dr. Ningwen Zhang Dr. Guusje Bonnema Laboratory of Plant Breeding, Wageningen University The Netherlands, Wageningen April-December, 2012 Investigating Hormone Regulation and Sugar Storage during Tuber Development in Turnip Plants (Brassica rapa) MSc Thesis Report (PBR-80436) Temesgen Menamo (Reg. No. 860717-557-050) Supervisors: Dr. Ningwen Zhang Dr. Guusje Bonnema Examiners : Dr. Guusje Bonnema Dr. Christian Bachem Laboratory of Plant Breeding, Wageningen University The Netherlands, Wageningen April-December, 2012 Acknowledgement I would like to thank the Brassica group leader Guusje Bonnema for giving me this opportunity to join in this group. I would like to thank Ningwen for support, guidance and encouragement in doing each part. I thank to Johan Bucher for his support in lab work. My thanks go to Luc Suurs who also supported me in biochemical lab works. I also would like to say thanks to Christian Bachem for being my examiner. I would like to thank for Fu Shi for helping on the transplanting of explants. I would like to acknowledge NFP for financially supporting until finishing of my study. Last but not least for all Brassica group members for support by ideas. Abstract Brassica rapa belongs to Brassicaceae family with diploid (2n=20) genotype and this species consists of morphological diverse crops. It includes leafy vegetables, turnip vegetables, and vegetable oil. This study focused on investigation of hormones and sugar storage during turnip tuber formation in turnip plants. -

Soybean Seedling Disease Management
SOYBEAN DISEASE MANAGEMENT CPN-1008 Soybean Seedling Diseases Soybean seedling diseases are one of the most important causes of reduced stand establishment and can cause economic losses. Diseased seedlings are often weak and less vigorous, and severe stand loss may require replanting. Seedling diseases are often more prevalent when wet weather follows planting. They are generally more common in compacted and poorly drained soils or in areas where seedlings have been injured (Figure 1). Diagnosing seedling diseases in the field can be difficult since the symptoms of various seedling blights are very similar. This publication discusses the characteristics of the organisms that cause seedling diseases, describes injury symptoms that may appear similar to these diseases, and briefly addresses how to manage seedling diseases. The soybean seedling diseases we examine are: Figure 1. Seedlings that die in patches or individually within a row are 1. Fusarium root rot symptoms of seedling blight diseases. 2. Rhizoctonia seedling blight 3. Phytophthora root rot 4. Pythium seedling blight 1 Soybean Seedling Diseases Fusarium Root Rot (Fusarium spp.) This disease can occur over a wide range of soil conditions. Many Fusarium species reside in the soil and can infect Affected plants typically appear in patches. Temperature soybean. Infected plants may be stunted and spindly, and and moisture requirements for infection vary, and stressed roots may have brown or black discoloration (Figure 2). seedlings may be more vulnerable to infection and disease. Affected plants may also have poorly developed root Phytophthora Root Rot systems. (Phytophthora sojae) In severe cases, seedlings may die before emerging. Phytophthora root rot is caused by the oomycete Fusarium species can infect plants under a wide variety Phytophthora sojae. -

THE ANATOMY and HISTOLOGY of the TRANSITION REGION of TRAGOPOGÓN PORRIFOLIUS ' by LEON HAVIS Assistant in Horticulture, Ohio Agricultural Experiment Station
THE ANATOMY AND HISTOLOGY OF THE TRANSITION REGION OF TRAGOPOGÓN PORRIFOLIUS ' By LEON HAVIS Assistant in horticulture, Ohio Agricultural Experiment Station INTRODUCTION Salsify {Tragopogón porrifolius L.) is a biennial plant belonging to one of the most specialized groups of the Compositae. During the first year of growth it produces a fleshy primary root, a fleshy hypo- cotyl, and a very short stem. In its second year a flower stalk is developed from the stem which extends to a height of 2 to 3 feet and bears disks of purple flowers. The seedling has linear, lanceolate, sessile cotyledons, which carry on photosynthesis. The fruit (com- monly called the seed) is a linear achene about 1 cm in length. REVIEW OF LITERATURE A search of the literature failed to reveal any reports dealing with the anatomy of the transition region of salsify. Van Tieghem (6) ^ in 1870 described very briefly the transition in a few plants of the Compositae, and Gérard (3) in 1881 reported the results of studies on the seedling anatomy of two members of the Compositae. One of these, Carthamus tindoriuSj is very similar in its type of transition to Tragopogón porrifolius in most details mentioned relative to the root and lower hypocotyl. Vuillemin (8) reported studies with the stems of some of the composites and included very brief notes on the transition phenomena. Dangeard (2) made very general obser- vations on types of transition and based the results of his final classi- fication on the number of primary xylem groups in the root. Van Tieghem (7, p. 782) defined three general types of transition from root to stem. -

Low Temperatures Affect Phase Transitions of Allium Sativum
agronomy Article From Embryo to Adult: Low Temperatures Affect Phase Transitions of Allium sativum L. from Germination to Flowering Tomer E. Ben Michael 1,2, Liraz Rozenblat 1,2, Adi Faigenboim 1, Einat Shemesh-Mayer 1, Itzhak Forer 1, Ross Peters 1, Joshua D. Klein 1, Haim D. Rabinowitch 2 and Rina Kamenetsky Goldstein 1,* 1 Institute of Plant Sciences, ARO, The Volcani Center, POB 6, Rishon LeZion 7505101, Israel; [email protected] (T.E.B.M.); [email protected] (L.R.); [email protected] (A.F.); [email protected] (E.S.-M.); [email protected] (I.F.); [email protected] (R.P.); [email protected] (J.D.K.) 2 Robert H. Smith Faculty of Agricultural, Food, and Environmental Quality Sciences, The Hebrew University of Jerusalem, Rehovot 76100, Israel; [email protected] * Correspondence: [email protected] Received: 3 October 2020; Accepted: 22 October 2020; Published: 26 October 2020 Abstract: Juvenile and vegetative adult shoot apical meristems (SAM) are actively involved in acquisition of flowering competence, while the embryonic SAM is regarded as “responsible” only for germination. Comparative analyses of imbibed and stratified seeds of garlic Allium sativum show that only stratified seedlings produced bulbs and flower stems at the end of the season. Since the seed morphology of stratified and non-stratified seeds prior to sowing was similar, the differences are attributed to the molecular alterations in the embryonic SAM. Functional annotation analysis of 3000 differentially expressed genes suggests that seed imbibition reactivates the embryonic cell cycle, initiates the metabolism, and primes garlic seed germination.