A Novel Widespread Bacterial Structure Related to the Flagellar Type III
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

C O N F E R E N C E 22 27 April 2016
Joint Pathology Center Veterinary Pathology Services WEDNESDAY SLIDE CONFERENCE 2015-2016 C o n f e r e n c e 22 27 April 2016 Cory Brayton, DVM, Ph.D., DACVP Associate Professor, Molecular & Comparative Pathobiology Johns Hopkins University School of Medicine Broadway Research Building, Suite 851 733 North Broadway Baltimore, MD 21205 CASE I: NIEHS-087 (JPC 4017222). Signalment: 11-month-old B6.129S- Cybbtm1Din/J mouse (Mus musculus) History: A breeding colony of B6.129S- Cybbtm1Din/J mice were housed in an AAALAC International accredited facility. The mice were housed in static micro isolator cases with ad libitum autoclaved food (NIH-31) and beta chip bedding. Mice were provided acidified water due to imm- unocompromised state. The mice were Body as a while, mouse. The liver was slightly enlarged, housed in the same room as B6 imm- and there are multiple tan foci in the liver and lung. (Photo courtesy of: National Institute of Environmental unocompetent mice. Sudden deaths were Health Sciences, Cellular and Molecular Pathology noted in the colony over a weekend. A total Branch and Comparative Medicine Branch, P.O. Box of 87 mice, aged from one to eleven months 12233, Research Triangle Park, NC 27709, http://www.niehs.nih.gov/research/atniehs/labs/lep/index. were affected. Of these, 45 mice were found cfm) dead and 19 sick mice were euthanized and were multifocal tan foci in the liver, spleen necropsied. Twenty males and 38 females and lung. were affected. Laboratory Results: From multiple tissues, Gross Pathology: The livers were pale and a pure culture of Burkholderia spp. -

Evaluation of the Relatedness of Brucella Spp. and Ochrobactrum Anthropi and Description of Ochrobactrum Intermedium Sp
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Dadun, University of Navarra International Journal of Systematic Bacteriology (1 998), 48, 759-768 Printed in Great Britain Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella SPP. Julian Velasco,’ Conchi Romero,’ lgnacio Lopez-Got%,’ Jose Leiva,2 Ramon Diaz1f2and lgnacio Moriydn’ Author for correspondence : Ignacio Moriyon. Tel : + 34 48 425600. Fax : + 34 48 425649. e-mail : [email protected] Departamento de The relatedness of Brucella spp. and Ochrobactrum anthropi was studied by M icrob io I og ia, Un ive rs id ad protein profiling, Western blot, immunoelectrophoresis and 16s rRNA analysis. de Navarra, Aptdo 1771 and Servicio de Microbiologia, Whole-cell and soluble proteins of brucellae and 0. anthropi showed Clinica Universitaria de serological cross-reactivities quantitatively and qualitatively more intense Navarraz, Pamplona, Spain than those existing with similar extracts of Agrobacterium spp. Numerical analysis of Western blot profiles of whole-cell extracts showed that 0. anthropi LMG 3301 was closer to Brucella spp. than to 0. anthropi LMG 3331T, a result not obtained by protein profiling. These differences were not observed by Western blot with soluble fractions, and immunoelectrophoretic analyses suggested that this was due to destruction of conformational epitopes in Western blot procedures with the subsequent simplification of antigenic profile. Analysis of the 165 rRNA sequences of strains previously used in the species definition confirmed that strain LMG 3301, and also LMG 3306, were closer to the brucellae, and that LMG 3331Twas in a separate cluster. -

Helicobacter Hepaticus Model of Infection: the Human Hepatocellular Carcinoma Controversy
Review articles Helicobacter hepaticus model of infection: the human hepatocellular carcinoma controversy Yessica Agudelo Zapata, MD,1 Rodrigo Castaño Llano, MD,2 Mauricio Corredor, PhD.3 1 Medical Doctor and General Surgeon in the Abstract Gastrohepatology Group at Universidad de Antioquia in Medellin, Colombia The discovery of Helicobacter 30 years ago by Marshall and Warren completely changed thought about peptic 2 Gastrointestinal Surgeon and Endoscopist in the and duodenal ulcers. The previous paradigm posited the impossibility of the survival of microorganisms in the Gastrohepatology Group at the Universidad de stomach’s low pH environment and that, if any microorganisms survived, they would stay in the duodenum Antioquia in Medellin, Colombia 3 Institute Professor of Biology in the Faculty of or elsewhere in the intestine. Today the role of H. pylori in carcinogenesis is indisputable, but little is known Natural Sciences and the GEBIOMIC Group the about other emerging species of the genus Helicobacter in humans. Helicobacter hepaticus is one of these Gastroenterology Group at the Universidad de species that has been studied most, after H. pylori. We now know about their microbiological, genetic and Antioquia in Medellin, Colombia pathogenic relationships with HCC in murine and human infections. This review aims to show the medical and ......................................... scientifi c community the existence of new species of Helicobacter that have pathogenic potential in humans, Received: 07-05-13 thus encouraging research. Accepted: 27-08-13 Keywords Helicobacter hepaticus, Helicobacter pylori, Helicobacter spp., hepatocellular carcinoma. INTRODUCTION bacterium can infect animals including mice, dogs and ger- bils which could lead to a proposal to use H. -

Iron Transport Strategies of the Genus Burkholderia
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2015 Iron transport strategies of the genus Burkholderia Mathew, Anugraha Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-113412 Dissertation Published Version Originally published at: Mathew, Anugraha. Iron transport strategies of the genus Burkholderia. 2015, University of Zurich, Faculty of Science. Iron transport strategies of the genus Burkholderia Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat.) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultät der Universität Zürich von Anugraha Mathew aus Indien Promotionskomitee Prof. Dr. Leo Eberl (Vorsitz) Prof. Dr. Jakob Pernthaler Dr. Aurelien carlier Zürich, 2015 2 Table of Contents Summary .............................................................................................................. 7 Zusammenfassung ................................................................................................ 9 Abbreviations ..................................................................................................... 11 Chapter 1: Introduction ....................................................................................... 14 1.1.Role and properties of iron in bacteria ...................................................................... 14 1.2.Iron transport mechanisms in bacteria ..................................................................... -

Rapid Identification of Klebsiella Pneumoniae, Corynebacterium Kutscheri, and Streptococcus Pneumoniae Using Triplex Polymerase Chain Reaction in Rodents
Exp. Anim. 62(1), 35–40, 2013 —Original— Rapid Identification of Klebsiella pneumoniae, Corynebacterium kutscheri, and Streptococcus pneumoniae Using Triplex Polymerase Chain Reaction in Rodents Eui-Suk JEONG1), Kyoung-Sun Lee1,2), Seung-Ho HEO1,3), Jin-Hee SEO1), and Yang-Kyu CHOI1) 1)Department of Laboratory Animal Medicine, College of Veterinary Medicine, Konkuk University, 1 Hwayang-dong, Gwangjin-gu, Seoul 143-701, Republic of Korea 2)Osong Medical Innovation Foundation Laboratory Animal Center, 186 Osong Saengmyung-Ro, Gangoe-myeon, Cheongwon-gun, Chungbuk 363-951, Republic of Korea 3)Asan Institute for Life Sciences, University of Ulsan College of Medicine, 388–1 Pungnap-2-dong, Songpa-gu Seoul 138-736, Republic of Korea Abstract: Klebsiella pneumoniae, Corynebacterium kutscheri, and Streptococcus pneumoniae are important pathogens that cause respiratory infections in laboratory rodents. In this study, we used species-specific triplex PCR analysis to directly detect three common bacterial pathogens associated with respiratory diseases. Specific targets were amplified with conventional PCR using thetyrB gene from K. pneumoniae, gyrB gene from C. kutscheri, and ply gene from S. pneumoniae. Our primers were tested against purified DNA from another eleven murine bacteria to determine primer specificity. Under optimal PCR conditions, the triplex assay simultaneously yielded a 931 bp product from K. pneumoniae, a 540 bp product from C. kutscheri, and a 354 bp product from S. pneumoniae. The triplex assay detection thresholds for pure cultures were 10 pg for K. pneumoniae and S. pneumoniae, and 100 pg for C. kutscheri. All three bacteria were successfully identified in the trachea and lung of experimentally infected mice at the same time. -

Endogenous Enterobacteriaceae Underlie Variation in Susceptibility to Salmonella Infection
ARTICLES https://doi.org/10.1038/s41564-019-0407-8 Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection Eric M. Velazquez1, Henry Nguyen1, Keaton T. Heasley1, Cheng H. Saechao1, Lindsey M. Gil1, Andrew W. L. Rogers1, Brittany M. Miller1, Matthew R. Rolston1, Christopher A. Lopez1,2, Yael Litvak 1, Megan J. Liou1, Franziska Faber 1,3, Denise N. Bronner1, Connor R. Tiffany1, Mariana X. Byndloss1,2, Austin J. Byndloss1 and Andreas J. Bäumler 1* Lack of reproducibility is a prominent problem in biomedical research. An important source of variation in animal experiments is the microbiome, but little is known about specific changes in the microbiota composition that cause phenotypic differences. Here, we show that genetically similar laboratory mice obtained from four different commercial vendors exhibited marked phenotypic variation in their susceptibility to Salmonella infection. Faecal microbiota transplant into germ-free mice repli- cated donor susceptibility, revealing that variability was due to changes in the gut microbiota composition. Co-housing of mice only partially transferred protection against Salmonella infection, suggesting that minority species within the gut microbiota might confer this trait. Consistent with this idea, we identified endogenous Enterobacteriaceae, a low-abundance taxon, as a keystone species responsible for variation in the susceptibility to Salmonella infection. Protection conferred by endogenous Enterobacteriaceae could be modelled by inoculating mice with probiotic Escherichia coli, which conferred resistance by using its aerobic metabolism to compete with Salmonella for resources. We conclude that a mechanistic understanding of phenotypic variation can accelerate development of strategies for enhancing the reproducibility of animal experiments. recent survey suggests that the majority of researchers Results have tried and failed to reproduce their own experiments To determine whether genetically similar strains of mice obtained A or experiments from other scientists1. -

Alvinella Pompejana Is an Endemic Inhabitant Tof Deep-Sea Hydrothermal Vents Located from 21°N to 32°S Latitude on the East Pacific Rise (1)
Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility Joseph J. Grzymskia,1, Alison E. Murraya,1, Barbara J. Campbellb, Mihailo Kaplarevicc, Guang R. Gaoc,d, Charles Leee, Roy Daniele, Amir Ghadirif, Robert A. Feldmanf, and Stephen C. Caryb,d,2 aDivision of Earth and Ecosystem Sciences, Desert Research Institute, 2215 Raggio Parkway, Reno, NV 89512; bCollege of Marine and Earth Studies, University of Delaware, Lewes, DE 19958; cDelaware Biotechnology Institute, 15 Innovation Way, Newark, DE 19702; dElectrical and Computer Engineering, University of Delaware, 140 Evans Hall, Newark, DE 19716; eDepartment of Biological Sciences, University of Waikato, Hamilton, New Zealand; fSymBio Corporation, 1455 Adams Drive, Menlo Park, CA 94025 Edited by George N. Somero, Stanford University, Pacific Grove, CA, and approved September 17, 2008 (received for review March 20, 2008) Hydrothermal vent ecosystems support diverse life forms, many of the thermal tolerance of a structural protein biomarker (5) which rely on symbiotic associations to perform functions integral supports the assertion that A. pompejana is likely among the to survival in these extreme physicochemical environments. Epsi- most thermotolerant and eurythermal metazoans on Earth lonproteobacteria, found free-living and in intimate associations (6, 7). with vent invertebrates, are the predominant vent-associated A. pompejana is characterized by a filamentous microflora that microorganisms. The vent-associated polychaete worm, Alvinella forms cohesive hair-like projections from mucous glands lining pompejana, is host to a visibly dense fleece of episymbionts on its the polychaete’s dorsal intersegmentary spaces (8). The episym- dorsal surface. The episymbionts are a multispecies consortium of biont community is constrained to the bacterial subdivision, Epsilonproteobacteria present as a biofilm. -

Appendix a Bacteria
Appendix A Complete list of 594 pathogens identified in canines categorized by the following taxonomical groups: bacteria, ectoparasites, fungi, helminths, protozoa, rickettsia and viruses. Pathogens categorized as zoonotic/sapronotic/anthroponotic have been bolded; sapronoses are specifically denoted by a ❖. If the dog is involved in transmission, maintenance or detection of the pathogen it has been further underlined. Of these, if the pathogen is reported in dogs in Canada (Tier 1) it has been denoted by an *. If the pathogen is reported in Canada but canine-specific reports are lacking (Tier 2) it is marked with a C (see also Appendix C). Finally, if the pathogen has the potential to occur in Canada (Tier 3) it is marked by a D (see also Appendix D). Bacteria Brachyspira canis Enterococcus casseliflavus Acholeplasma laidlawii Brachyspira intermedia Enterococcus faecalis C Acinetobacter baumannii Brachyspira pilosicoli C Enterococcus faecium* Actinobacillus Brachyspira pulli Enterococcus gallinarum C C Brevibacterium spp. Enterococcus hirae actinomycetemcomitans D Actinobacillus lignieresii Brucella abortus Enterococcus malodoratus Actinomyces bovis Brucella canis* Enterococcus spp.* Actinomyces bowdenii Brucella suis Erysipelothrix rhusiopathiae C Actinomyces canis Burkholderia mallei Erysipelothrix tonsillarum Actinomyces catuli Burkholderia pseudomallei❖ serovar 7 Actinomyces coleocanis Campylobacter coli* Escherichia coli (EHEC, EPEC, Actinomyces hordeovulneris Campylobacter gracilis AIEC, UPEC, NTEC, Actinomyces hyovaginalis Campylobacter -
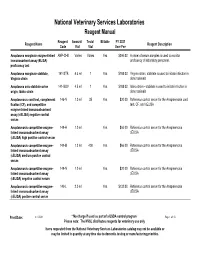
NVSL Reagent Manual
National Veterinary Services Laboratories Reagent Manual Reagent Amount/ Tests/ Billable FY 2021 Reagent Name Reagent Description Code Vial Vial User Fee Anaplasma marginale enzyme-linked ANP-CHK Varies VariesYes $394.00 A panel of serum samples is used to monitor immunosorbent assay (ELISA) proficiency of laboratory personnel. proficiency test Anaplasma marginale stabilate, 141-STB 4.5 ml 1Yes $188.00 Virginia strain, stabilate is used to initiate infection in Virginia strain donor animals Anaplasma ovis stabilate ovine 141-SOV 4.5 ml 1Yes $188.00 Idaho strain - stabilate is used to initate infection in origin, Idaho strain donor animals Anaplasmosis card test, complement 146-N 1.0 ml 35Yes $20.00 Reference control serum for the Anaplasmosis card fixation (CF), and competitive test, CF, and cELISA enzyme-linked immunoabsorbent assay (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-H 1.0 mlYes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) high positive control serum Anaplasmosis competitive enzyme- 149-M 1.0 ml 400Yes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) medium positve control serum Anaplasmosis competitive enzyme- 149-N 1.0 mlYes $20.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-L 2.0 mlYes $132.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) positve control serum Print Date: 6/11/2021 * No charge if used as part of a USDA control program Page 1 of 58 Please note: The NVSL distributes reagents for veterinary use only Items requested from the National Veterinary Services Laboratories catalog may not be available or may be limited in quantity at any time due to domestic testing or manufacturing priorities. -

APUTS) Reporting Terminology and Codes Microbiology (V1.0
AUSTRALIAN PATHOLOGY UNITS AND TERMINOLOGY (APUTS) Reporting Terminology and Codes Microbiology (v1.0) 1 12/02/2013 APUTS Report Information Model - Urine Microbiology Page 1 of 1 Specimen Type Specimen Macro Time Glucose Bilirubin Ketones Specific Gravity pH Chemistry Protein Urobilinogen Nitrites Haemoglobin Leucocyte Esterases White blood cell count Red blood cells Cells Epithelial cells Bacteria Microscopy Parasites Microorganisms Yeasts Casts Crystals Other elements Antibacterial Activity No growth Mixed growth Urine MCS No significant growth Klebsiella sp. Bacteria ESBL Klebsiella pneumoniae Identification Virus Fungi Growth of >10^8 org/L 10^7 to 10^8 organism/L of mixed Range or number Colony Count growth of 3 organisms 19090-0 Culture Organism 1 630-4 LOINC >10^8 organisms/L LOINC Significant growth e.g. Ampicillin 18864-9 LOINC Antibiotics Susceptibility Method Released/suppressed None Organism 2 Organism 3 Organism 4 None Consistent with UTI Probable contamination Growth unlikely to be significant Comment Please submit a repeat specimen for testing if clinically indicated Catheter comments Sterile pyuria Notification to infection control and public health departments PUTS Urine Microbiology Information Model v1.mmap - 12/02/2013 - Mindjet 12/02/2013 APUTS Report Terminology and Codes - Microbiology - Urine Page 1 of 3 RCPA Pathology Units and Terminology Standardisation Project - Terminology for Reporting Pathology: Microbiology : Urine Microbiology Report v1 LOINC LOINC LOINC LOINC LOINC LOINC LOINC Urine Microbiology Report -

Characterizing the Abcr/Vtlr System in the Rhizobiales
Characterizing the AbcR/VtlR system in the Rhizobiales Lauren Marie Sheehan Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical and Veterinary Sciences Clayton C. Caswell, Chair Thomas J. Inzana Birgit Scharf Nammalwar Sriranganathan April 20, 2018 Blacksburg, Virginia Keywords: Brucella, LysR-type transcriptional regulators, small RNAs Characterizing the AbcR/VtlR system in the Rhizobiales Lauren Marie Sheehan Abstract Rhizobiales encompass a diverse group of microbes, ranging from free-living, soil- dwelling bacteria to disease-causing, intracellular pathogens. Although the lifestyle of these organisms vary, many genetic systems are well conserved. One system, named the AbcR/VtlR system, is found throughout rhizobiales, and even extends to bacteria in other orders within the Alphaproteobacteria. The AbcR sRNAs are an example of sibling sRNAs, where two copies of the abcR gene are typically present in the genome. The AbcRs are involved in the negative regulation of ABC-type transport systems, which are important components for nutrient acquisition. Although the AbcRs share several features amongst organisms, major differences can be found in their functional and regulatory redundancy, the targets they regulate and how they regulate them. Specifically, one major difference in the AbcRs lies in the nucleotide sequences utilized by the sRNAs to bind mRNA targets. In the present studies, the regulatory mechanisms of the AbcR sRNAs were further characterized in the mammalian pathogen Brucella abortus, and the full regulatory profiles of the AbcRs were defined in the plant pathogen Agrobacterium tumefaciens. As mentioned above, the AbcR sRNAs are important for the proper regulation of nutrient-acquiring transport systems in the Rhizobiales. -

Tese Adelina Margarida Parente
Defences of Helicobacter species against host antimicrobials Adelina Margarida Lima Pereira Rodrigues Parente Dissertation presented to obtain the Ph.D degree in Biochemistry Instituto de Tecnologia Química e Biológica António Xavier | Universidade Nova de Lisboa Oeiras, June, 2016 Defences of Helicobacter species against host antimicrobials Adelina Margarida Lima Pereira Rodrigues Parente Dissertation presented to obtain the Ph.D degree in Biochemistry Instituto de Tecnologia Química e Biológica António Xavier | Universidade Nova de Lisboa Oeiras, June 2016 From left to right: Mónica Oleastro (4th oponente), Gabriel Martins (3 rd opponent), Marta Justino (Co-supervisor) , Miguel Viveiros (2 nd opponent), Adelina Margarida Parente , Lígia Saraiva (supervisor) , Cecília Arraiano (president of the jury), and Maria do Céu Figueiredo (1 st opponent). nd 22 June 2016 Second edition, June 2016 Molecular Mechanisms of Pathogen Resistance Laboratory Instituto de Tecnologia Química e Biológica António Xavier Universidade Nova de Lisboa 2780-157 Portugal ii “Science knows no country, because knowledge belongs to humanity, and is the torch which illuminates the world” Louis Pasteur iii iv Acknowledgments Firstly, I would like to express my gratitude to the person that allowed me the opportunity to perform a PhD and who also contributed the most for my accomplishment of this thesis by constantly supporting me during these last years. I thus thank my supervisor, Dr. Lígia Saraiva , for her permanent availability whenever I needed guidance, for all the excellent ideas and advices related to my practical work and lastly for all the patience and enthusiasm! I also thank Dr. Lígia for her rigour and enormous help in the writing of this thesis.