Hymenoptera: Vespidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mitochondrial Genome of the Stonefly Kamimuria Wangi (Plecoptera: Perlidae) and Phylogenetic Position of Plecoptera Based on Mitogenomes
Mitochondrial Genome of the Stonefly Kamimuria wangi (Plecoptera: Perlidae) and Phylogenetic Position of Plecoptera Based on Mitogenomes Qian Yu-Han1,2, Wu Hai-Yan1, Ji Xiao-Yu1, Yu Wei-Wei1, Du Yu-Zhou1* 1 School of Horticulture and Plant Protection and Institute of Applied Entomology, Yangzhou University, Yangzhou, Jiangsu, China, 2 College of Forestry, Southwest Forestry University, Kunming, Yunnan, China Abstract This study determined the mitochondrial genome sequence of the stonefly, Kamimuria wangi. In order to investigate the relatedness of stonefly to other members of Neoptera, a phylogenetic analysis was undertaken based on 13 protein-coding genes of mitochondrial genomes in 13 representative insects. The mitochondrial genome of the stonefly is a circular molecule consisting of 16,179 nucleotides and contains the 37 genes typically found in other insects. A 10-bp poly-T stretch was observed in the A+T-rich region of the K. wangi mitochondrial genome. Downstream of the poly-T stretch, two regions were located with potential ability to form stem-loop structures; these were designated stem-loop 1 (positions 15848– 15651) and stem-loop 2 (15965–15998). The arrangement of genes and nucleotide composition of the K. wangi mitogenome are similar to those in Pteronarcys princeps, suggesting a conserved genome evolution within the Plecoptera. Phylogenetic analysis using maximum likelihood and Bayesian inference of 13 protein-coding genes supported a novel relationship between the Plecoptera and Ephemeroptera. The results contradict the existence of a monophyletic Plectoptera and Plecoptera as sister taxa to Embiidina, and thus requires further analyses with additional mitogenome sampling at the base of the Neoptera. -
The Mitochondrial Genomes of Palaeopteran Insects and Insights
www.nature.com/scientificreports OPEN The mitochondrial genomes of palaeopteran insects and insights into the early insect relationships Nan Song1*, Xinxin Li1, Xinming Yin1, Xinghao Li1, Jian Yin2 & Pengliang Pan2 Phylogenetic relationships of basal insects remain a matter of discussion. In particular, the relationships among Ephemeroptera, Odonata and Neoptera are the focus of debate. In this study, we used a next-generation sequencing approach to reconstruct new mitochondrial genomes (mitogenomes) from 18 species of basal insects, including six representatives of Ephemeroptera and 11 of Odonata, plus one species belonging to Zygentoma. We then compared the structures of the newly sequenced mitogenomes. A tRNA gene cluster of IMQM was found in three ephemeropteran species, which may serve as a potential synapomorphy for the family Heptageniidae. Combined with published insect mitogenome sequences, we constructed a data matrix with all 37 mitochondrial genes of 85 taxa, which had a sampling concentrating on the palaeopteran lineages. Phylogenetic analyses were performed based on various data coding schemes, using maximum likelihood and Bayesian inferences under diferent models of sequence evolution. Our results generally recovered Zygentoma as a monophyletic group, which formed a sister group to Pterygota. This confrmed the relatively primitive position of Zygentoma to Ephemeroptera, Odonata and Neoptera. Analyses using site-heterogeneous CAT-GTR model strongly supported the Palaeoptera clade, with the monophyletic Ephemeroptera being sister to the monophyletic Odonata. In addition, a sister group relationship between Palaeoptera and Neoptera was supported by the current mitogenomic data. Te acquisition of wings and of ability of fight contribute to the success of insects in the planet. -
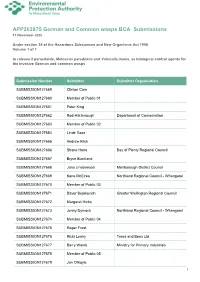
APP203875 Submissions Compilation.Pdf
APP203875 German and Common wasps BCA Submissions 11 November 2020 Under section 34 of the Hazardous Substances and New Organisms Act 1996 Volume 1 of 1 to release 2 parasitoids, Metoecus paradoxus and Volucella inanis, as biological control agents for the invasive German and common wasps Submission Number Submitter Submitter Organisation SUBMISSION127659 Clinton Care SUBMISSION127660 Member of Public 01 SUBMISSION127661 Peter King SUBMISSION127662 Rod Hitchmough Department of Conservation SUBMISSION127663 Member of Public 02 SUBMISSION127664 Linde Rose SUBMISSION127665 Andrew Blick SUBMISSION127666 Shane Hona Bay of Plenty Regional Council SUBMISSION127667 Bryce Buckland SUBMISSION127668 Jono Underwood Marlborough District Council SUBMISSION127669 Kane McElrea Northland Regional Council - Whangarei SUBMISSION127670 Member of Public 03 SUBMISSION127671 Davor Bejakovich Greater Wellington Regional Council SUBMISSION127672 Margaret Hicks SUBMISSION127673 Jenny Dymock Northland Regional Council - Whangarei SUBMISSION127674 Member of Public 04 SUBMISSION127675 Roger Frost SUBMISSION127676 Ricki Leahy Trees and Bees Ltd SUBMISSION127677 Barry Wards Ministry for Primary Industries SUBMISSION127678 Member of Public 05 SUBMISSION127679 Jan O'Boyle 1 Submission Number Submitter Submitter Organisation Apiculture New Zealand Science and SUBMISSION127680 Sue Carter Research Focus Group SUBMISSION127681 Andrea Dorn SUBMISSION127682 Benita Wakefield Te Rūnanga o Ngāi Tahu SUBMISSION127683 Emma Edney-Browne Auckland Council SUBMISSION127684 David Hunter -

Monopoly of Reproduction by the Queen in the Social Wasp Ropalidia
Proc Indian Natn Sci Acad 80 No. 5 December 2014 pp. 1025-1044 Printed in India. DOI: 10.16943/ptinsa/2014/v80i5/47971 Review Article Queen Pheromone and Monopoly of Reproduction by the Queen in the Social Wasp Ropalidia marginata ANIRUDDHA MITRA Laboratoire Evolution, Génomes et Spéciation, Centre National de la Recherche Scientifique, avenue de la Terrasse, Batiment 13, Gif sur Yvette, 91190, France (Received on 29 April 2014; Revised on 30 September 2014; Accepted on 13 October 2014) Ropalidia marginata is a primitively eusocial (truly social) wasp found in peninsular India. It is different from the typical primitively eusocial species in having docile queens that cannot use aggression to maintain reproductive monopoly. Recent studies using chemical analysis and bioassays indicate that Dufour’s gland is a source of the queen pheromone in this species. Queens appear to signal their presence to workers through their Dufour’s gland compounds, possibly by applying them on the nest surface, and this results in suppression of reproduction by workers, resulting in reproductive monopoly by the queen. The Dufour’s gland was found to contain saturated long chain hydrocarbons, which have recently been suggested to be the ancestral state of fertility signals in Hymenoptera. The Dufour’s gland composition differed significantly between queens and workers, and was also correlated with the state of ovarian development, varying continuously as a function of ovarian development, thereby advocating the honesty of the queen pheromone. This elucidates the mechanism of maintenance of eusociality through pheromonal queen signalling by the Dufour’s gland compounds. Key Words: Eusocial; Reproductive Monopoly; Queen Pheromone; Dufour’s Gland; Ropalidia marginata; Honest Signal Social insects like bees, ants and wasps have aroused whole theory. -

Comparative Morphology of the Male Genitalia in Lepidoptera
COMPARATIVE MORPHOLOGY OF THE MALE GENITALIA IN LEPIDOPTERA. By DEV RAJ MEHTA, M. Sc.~ Ph. D. (Canta.b.), 'Univefsity Scholar of the Government of the Punjab, India (Department of Zoology, University of Oambridge). CONTENTS. PAGE. Introduction 197 Historical Review 199 Technique. 201 N ontenclature 201 Function • 205 Comparative Morphology 206 Conclusions in Phylogeny 257 Summary 261 Literature 1 262 INTRODUCTION. In the domains of both Morphology and Taxonomy the study' of Insect genitalia has evoked considerable interest during the past half century. Zander (1900, 1901, 1903) suggested a common structural plan for the genitalia in various orders of insects. This work stimulated further research and his conclusions were amplified by Crampton (1920) who homologized the different parts in the genitalia of Hymenoptera, Mecoptera, Neuroptera, Diptera, Trichoptera Lepidoptera, Hemiptera and Strepsiptera with those of more generalized insects like the Ephe meroptera and Thysanura. During this time the use of genitalic charac ters for taxonomic purposes was also realized particularly in cases where the other imaginal characters had failed to serve. In this con nection may be mentioned the work of Buchanan White (1876), Gosse (1883), Bethune Baker (1914), Pierce (1909, 1914, 1922) and others. Also, a comparative account of the genitalia, as a basis for the phylo genetic study of different insect orders, was employed by Walker (1919), Sharp and Muir (1912), Singh-Pruthi (1925) and Cole (1927), in Orthop tera, Coleoptera, Hemiptera and the Diptera respectively. It is sur prising, work of this nature having been found so useful in these groups, that an important order like the Lepidoptera should have escaped careful analysis at the hands of the morphologists. -

Polistes Dominula's Impact on P. Fuscatus in the Northeastern US
Biol Invasions https://doi.org/10.1007/s10530-017-1617-8 ORIGINAL PAPER Displacement and replacement in real time: Polistes dominula’s impact on P. fuscatus in the northeastern U.S. Julia A. Pilowsky . Philip T. Starks Received: 11 May 2017 / Accepted: 7 November 2017 Ó Springer International Publishing AG, part of Springer Nature 2017 Abstract Two major challenges in studying the at the most invaded sites. These findings suggest a impacts of exotic invasive species on native species positive feedback cycle in the establishment of P. are identifying mechanisms of displacement and dominula, in which the invasive wasp drives popula- replacement and the lack of long-term population tion declines in the native that in turn allow P. studies in these systems. A solution for the first is to dominula to further establish. This system provides an study invasive and native congeners that occupy the example of a possible extinction vortex caused by same niche. A solution for the second is to study many competitive exclusion of a species by its invasive populations for one year instead of one population for congener. many years. We studied the invasion biology of the invasive European paper wasp Polistes dominula and Keywords Polistes Á Invasion biology Á Competitive its native congener the Northern paper wasp P. exclusion Á Local extinction Á Displacement fuscatus, two species which compete for similar resources. We tracked the demography of the two wasps at sites in the northeastern United States. We found that the survival of P. dominula to the repro- Introduction ductive period in August was three times that of P. -
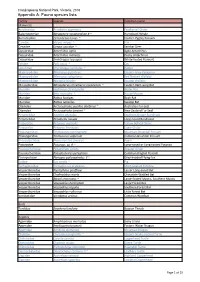
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus -

Biodiversiteitsopname Biodiversity Assessment
Biodiversiteitsopname Biodiversity Assessment Bome - Trees (77 sp) Veldblomme - Flowering veld plants (65 sp) Grasse - Grasses (41 sp) Naaldekokers - Dragonflies (46 sp) Skoenlappers - Butterflies (81 sp) Motte - Moths (95 sp) Nog insekte - Other insects (102 sp) Spinnekoppe - Spiders (53 sp) Paddas - Frogs (14 sp) Reptiele - Reptiles (22 sp) Voëls - Birds (185 sp) Soogdiere - Mammals (23 sp) 4de uitgawe: Jan 2015 Plante/Plants Diere/Animals (24 000 spp in SA) Anthropoda Chordata (>150 000 spp in SA) Arachnida Insecta (spinnekoppe/spiders, 2020 spp in SA) Neuroptera – mayflies, lacewings, ant-lions (385 spp in SA) Odonata – dragonflies (164 spp in SA) Blattodea – cockroaches (240 spp in SA) Mantodea – mantids (185 spp in SA) Isoptera – termites (200 spp in SA) Orthoptera – grasshoppers, stick insects (900 spp in SA) Phthiraptera – lice (1150 spp in SA) Hemiptera – bugs (>3500 spp in SA) Coleoptera – beetles (18 000 spp in SA) Lepidoptera – butterflies (794 spp in SA), moths (5200 spp in SA) Diptera – flies (4800 spp in SA) Siphonoptera – fleas (100 spp in SA) Hymenoptera – ants, bees, wasps (>6000 spp in SA) Trichoptera – caddisflies (195 spp in SA) Thysanoptera – thrips (230 spp in SA) Vertebrata Tunicata (sea creatures, etc) Fish Amphibia Reptiles Birds Mammals (115 spp in SA) (255 spp in SA) (858 spp in SA) (244 spp in SA) Bome – Trees (n=77) Koffiebauhinia - Bauhinia petersiana - Dainty bauhinia Rooi-ivoor - Berchemia zeyheri - Red ivory Witgat - Boscia albitrunca - Shepherd’s tree Bergvaalbos - Brachylaena rotundata - Mountain silver-oak -

A Survey on Sphingidae (Lepidoptera) Species of South Eastern Turkey
Cumhuriyet Science Journal e-ISSN: 2587-246X Cumhuriyet Sci. J., 41(1) (2020) 319-326 ISSN: 2587-2680 http://dx.doi.org/10.17776/csj.574903 A survey on sphingidae (lepidoptera) species of south eastern Turkey with new distributional records Erdem SEVEN 1 * 1 Department of Gastronomy and Culinary Arts, School of Tourism and Hotel Management, Batman University, 72060, Batman, Turkey. Abstract Article info History: This paper provides comments on the Sphingidae species of south eastern Turkey by the field Received:10.06.2019 surveys are conducted between in 2015-2017. A total of 15 species are determined as a result Accepted:20.12.2019 of the investigations from Batman, Diyarbakır and Mardin provinces. With this study, the Keywords: number of sphinx moths increased to 13 in Batman, 14 in Diyarbakır and 8 in Mardin. Among Fauna, them, 7 species for Batman, 4 species for Diyarbakır and 1 species for Mardin are new record. Hawk moths, For each species, original reference, type locality, material examined, distribution in the world New records, and in Turkey, and larval hostplants are given. Adults figures of Smerinthus kindermanni Sphingidae, Lederer, 1852; Marumba quercus ([Denis & Schiffermüller], 1775); Rethera komarovi Turkey. (Christoph, 1885); Macroglossum stellatarum (Linnaeus, 1758); Hyles euphorbiae (Linnaeus, 1758) and H. livornica (Esper, [1780]) are illustrated. 1. Introduction 18, 22-24]: Acherontia atropos (Linnaeus, 1758); Agrius convolvuli (Linnaeus, 1758); Akbesia davidi (Oberthür, 1884); Clarina kotschyi (Kollar, [1849]); C. The Sphingidae family classified in the Sphingoidea syriaca (Lederer, 1855); Daphnis nerii (Linnaeus, Superfamily and species of the family are generally 1758); Deilephila elpenor (Linnaeus, 1758); D. -

Exhibition Catalogue Natural History Illustrations by Erin Forsyth, 2018
A Few Exhibition catalogue Natural history illustrations by Erin Forsyth, 2018 TABLE OF CONTENTS ABOUT THE WORKS 5 About the artist 7 How to use this catalogue 9 TERMS AND CONDITIONS OF SALE 10 Korimako, makomako, bellbird 13 Kākāriki, Red-crowned parakeet, (Cyanoramphus novaezelandiae) 15 Moko kākāriki, Auckland green gecko (Naultinus elegans) 17 Pekapeka-tou-roa, long-tailed bat (Chalinolobus tuberculatus) 19 Pekapeka-tou-roa, long-tailed bat (Chalinolobus tuberculatus) 21 Pekapeka-tou-roa, long-tailed bat (Chalinolobus tuberculatus) 23 Ngirungiru, miromiro, South Island tomtit (Petroica macrocephala macrocephala) male 25 Kakaruwai, South Island Robin (Petroica australis) 27 Tōrea pango, variable oystercatcher (Haematopus unicolor) 29 Kererū, NZ wood pigeon (Hemiphaga novaeseelandiae) 31 Kōtare, sacred kingfisher (Todiramphus sanctus) 33 Ruru, morepork (Ninox novaeseelandiae) 35 TŪī, parsons bird (Prosthemadera novaeseelandiae) 37 Kōkako, blue-wattled crow (Callaeas wilsoni) 41 Takahe, South Island Takahe (Porphyrio hochstetteri) 43 Tūturiwhatu, NZ Dotteral (Charadrius obscurus) 45 Whio, blue duck (Hymenolaimus malacorhynchos) 47 Kahukōwhai, yellow admiral (Vanessa itea) 49 Wētāpunga, Little Barrier (Hauturu-o-Toi) giant weta (Deinacrida heteracantha) 51 Kārearea, NZ falcon (Falco novaeseelandiae) 53 Common evening brown (Melanitis leda bankia) 55 Pepe pouri, Helms' butterfly or forest ringlet (Dodonidia helmsii) 59 Kahukōwhai, yellow admiral (Vanessa itea) & Kahukura, NZ red admiral (V. gonerilla gonerilla) 63 Pepe pouri, Butler's ringlet (Erebiola butleri) & pepe pouri, black mountain ringlet (Percnodaimon merula) 67 Pīwakawaka, fantail (Rhipidura fuliginosa) 73 Weka, woodhen (Gallirallus australis) 75 Carnivorous land snail (Powelliphanta superba) 77 MYRTACEAE Studies I & II (Diptych) 79 ABOUT THE WORKS These original works are from the exhibition ‘A Few’ - the third installment in an ongoing series of natural history illustrations depicting native and resident species of Aotearoa by Erin Forsyth. -
Home Pre-Fire Moth Species List by Species
Species present before fire - by species Scientific Name Common Name Family Abantiades aphenges Hepialidae Abantiades hyalinatus Mustard Ghost Moth Hepialidae Abantiades labyrinthicus Hepialidae Acanthodela erythrosema Oecophoridae Acantholena siccella Oecophoridae Acatapaustus leucospila Nolidae Achyra affinitalis Cotton Web Spinner Crambidae Aeolochroma mniaria Geometridae Ageletha hemiteles Oecophoridae Aglaosoma variegata Notodontidae Agriophara discobola Depressariidae Agrotis munda Brown Cutworm Noctuidae Alapadna pauropis Erebidae Alophosoma emmelopis Erebidae Amata nigriceps Erebidae Amelora demistis Pointed Cape Moth Geometridae Amelora sp. Cape Moths Geometridae Antasia flavicapitata Geometridae Anthela acuta Common Anthelid Moth Anthelidae Anthela ferruginosa Anthelidae Anthela repleta Anthelidae Anthela sp. Anthelidae Anthela varia Variable Anthelid Anthelidae Antipterna sp. Oecophoridae Ardozyga mesochra Gelechiidae Ardozyga sp. Gelechiidae Ardozyga xuthias Gelechiidae Arhodia lasiocamparia Pink Arhodia Geometridae Arrade destituta Erebidae Arrade leucocosmalis Erebidae Asthenoptycha iriodes Tortricidae Asura lydia Erebidae Azelina biplaga Geometridae Barea codrella Oecophoridae Calathusa basicunea Nolidae Calathusa hypotherma Nolidae Capusa graodes Geometridae Capusa sp. Geometridae Carposina sp. Carposinidae Casbia farinalis Geometridae Casbia sp. Geometridae Casbia tanaoctena Geometridae Catacometes phanozona Oecophoridae Catoryctis subparallela Xyloryctidae Cernia amyclaria Geometridae Chaetolopha oxyntis Geometridae Chelepteryx -

º'‡ Èõàπõπ§ ∫ (Lepidoptera: Geometridae) ¢Õ߇¢Μ√—°…“Æ
π‘æπ∏åµâπ©∫—∫ º’‡ ◊ÈÕÀπÕπ§◊∫ (Lepidoptera: Geometridae) ¢Õ߇¢µ√—°…“æ—π∏ÿå —µ«åªÉ“ Œ“≈“-∫“≈“ ®—ßÀ«—¥π√“∏‘«“ »ÿ¿ƒ°…å «—≤π ‘∑∏‘Ï 1 ™—¬«—≤πå ª√–¡«≈2 ÿ√‰°√ ‡æ‘Ë¡§”3 ·≈– »‘√‘æ√ ∑ÕßÕ“√’¬å 4 Abstract Watanasit, S.1, Pramual, C.1, Permkam, S.2, and Thong-Aree, S.3 Geometrid moths (Lepidoptera: Geometridae) in Hala-Bala Wildlife Sanctuary, Narathiwat Province Songklanakarin J. Sci. Technol., 2004, 26(2) : 197-210 The purpose of this research was to investigate the species diversity and abundance of geometrid moths in tropical rain forest of Hala-Bala Wildlife Sanctuary (< 200 meters above sea level), Narathiwat Province, southern Thailand. Field data were collected every 2 months from July 2001 to July 2002. Three light traps were placed 200 meters apart. Moths were collected every 2 hours between 18.00 - 24.00 pm for 1Department of Biology, Faculty of Science, 2Department of Pest Management, Faculty of Natural Resourecs, Prince of Songkla University, Hat Yai, Songkhla, 90112, 3Peat Swamp Hala-Bala Research Station, Narathiwat, 96160 Thailand. 1«∑.¡. ( —µ««‘∑¬“) √Õß»“ µ√“®“√¬å 2«∑.¡. (𑇫»«‘∑¬“) π—°»÷°…“ ¿“§«‘™“™’««‘∑¬“ §≥–«‘∑¬“»“ µ√å 3Ph.D. (Entomology) √Õß»“ µ√“®“√¬å ¿“§«‘™“°“√®—¥°“√»—µ√Ÿæ◊™ §≥–∑√—欓°√∏√√¡™“µ‘ ¡À“«‘∑¬“≈—¬ ߢ≈“π§√‘π∑√å Õ”‡¿ÕÀ“¥„À≠à ®—ßÀ«—¥ ߢ≈“ 90112 4«∑.¡. (™’««‘∑¬“ªÉ“‰¡â) π—°«‘™“°“√ ∂“π’«‘®—¬ —µ«åªÉ“ ªÉ“æ√ÿªÉ“Œ“≈“-∫“≈“ 96160 π√“∏‘«“ Corresponding e-mail : [email protected] √—∫µâπ©∫—∫ 10 µÿ≈“§¡ 2546 √—∫≈ßæ‘¡æå 18 µÿ≈“§¡ 2546 Songklanakarin J. Sci. Technol. Geometrid moths in Hala-Bala Wildlife Sanctuary, Narathiwat Vol. 26 No. 2 Mar.-Apr. 2004 198 Watanasit, S., et al. 3 consecutive nights. Seven hundred and fifty six individuals of geometrid moths comprising 5 subfamilies, 17 tribes, 67 genera and 129 species were collected and identified.