Hynobiid Salamanders (Paradactylodon Persicus S.L.) from Iran, Illuminated by Phylogeographic, Developmental, and Transcriptomic Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Resolution, Multi-Proxy Peat Record from NW Iran: the Hand That Rocked the Cradle of Civilization?
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/280075839 Abrupt climate variability since the last deglaciation based on a high- resolution, multi-proxy peat record from NW Iran: The hand that rocked the Cradle of Civilization? Article in Quaternary Science Reviews · August 2015 DOI: 10.1016/j.quascirev.2015.07.006 CITATIONS READS 41 1,081 12 authors, including: Arash Sharifi Ali Pourmand University of Miami University of Miami 74 PUBLICATIONS 166 CITATIONS 75 PUBLICATIONS 1,062 CITATIONS SEE PROFILE SEE PROFILE Elizabeth Canuel Larry C. Peterson Virginia Institute of Marine Science University of Miami 131 PUBLICATIONS 5,297 CITATIONS 176 PUBLICATIONS 10,501 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Madison Group Project View project ANR-PALEOPERSEPOLIS View project All content following this page was uploaded by Ali Pourmand on 10 September 2018. The user has requested enhancement of the downloaded file. Quaternary Science Reviews 123 (2015) 215e230 Contents lists available at ScienceDirect Quaternary Science Reviews journal homepage: www.elsevier.com/locate/quascirev Abrupt climate variability since the last deglaciation based on a high-resolution, multi-proxy peat record from NW Iran: The hand that rocked the Cradle of Civilization? * Arash Sharifi a, b, , Ali Pourmand a, b, Elizabeth A. Canuel c, Erin Ferer-Tyler c, Larry C. Peterson b, Bernhard Aichner d, Sarah J. Feakins d, Touraj Daryaee e, Morteza Djamali f, Abdolmajid Naderi Beni g, Hamid A.K. Lahijani g, Peter K. Swart b a Neptune Isotope Laboratory (NIL), Department of Marine Geosciences, Rosenstiel School of Marine and Atmospheric Science, University of Miami, 4600 Rickenbacker Causeway, Miami, FL 33149-1098, USA b Department of Marine Geosciences, Rosenstiel School of Marine and Atmospheric Science (RSMAS), University of Miami, 4600 Rickenbacker Causeway, Miami, FL 33149-1098, USA c Virginia Institute of Marine Science, College of William & Mary, P.O. -

LARVAL FORMS in ECHINODERMATA in Echinoderms
LARVAL FORMS IN ECHINODERMATA In echinoderms eggs and sperms are released in water and fertilization takes place in water forming zygote. Echinoderms are deuterostomes and hence cleavage is radial, holoblastic and indeterminate. The larvae hatch in water and feed and grow through successive larval stages to become adults. The larvae of echinoderms are bilaterally symmetrical but lose symmetry during metamorphosis. Different classes of echinoderms show structurally different larval stages and their comparisons can reveal their evolutionary ancestry. LARVAE OF ASTEROIDEA There are three larval stages in Asteroidea in the course of their development to adult stage. Early bipinnaria appears like hypothetical dipleurula. It has oval body without arms and ciliary bands for locomotion. It has well developed alimentary canal for feeding and grows to become bipinnaria. Bipinnaria larva possesses 5 pairs of ciliated arms which do not have any skeletal support inside. These arms are used for swimming in water while feeding on planktons. Preoral and postoral ciliary bands are also present. This larva resembles auricularia larva of Holothuroidea in general appearance. Brachiolaria larva is formed after 6-7 weeks of life and growth of bipinnaria. This larva is sedentary and remains attached to a hard substratum for which it possesses three brachiolarian arms having adhesive discs at the tip. Ciliated arms get reduced and become thin and functionless, while mouth, anus and gut are well developed. It has axocoel, hydocoel and somatocoel that later on give rise to water vascular system Development of starfish takes place inside the sedentary brachiolaria which ruptures and releases tiny starfish into water. LARVAE OF HOLOTHUROIDEA Class Holothuroidea demonstrate two larval stages, namely, auricularia and doliolaria larvae. -

First Horseback Azerbaidjan Expedition 2018 Indiv
Pioneering Azerbaïdjan Horse Expedition CAUCASUS EXPEDITIONS Contact : Audrey Bogini [email protected] [email protected] CAUCASUS EXPEDITIONS 2018 Azerbaidjan shares its border with Georgia, Dagestan, Iran and Armenia. It stretches from the North with the Great Caucasus Mountains to the hilly picturesque Hirkan Natio- nal Park and the Talysh mountains in the South. It is the only country of the whole Cauca- sus chain where you can cross the southern slopes to the northern slopes. In the Caucasus area, Azerbaidjan is nowadays the wildest country for nature activities. Focused on the oil business the decision makers were not thinking about developing sport / outdoors tourism. The core of our itinerary is the self sufficient horseback trek in the very wilderness of the Great Caucasus Mountains. During our route we ride over passes, on crest or high pla- teau, between mountains lakes, through untouched forest and wild rivers. We will be accompanied by the local Lezgian people who know by heart the incredible Ismaili Na- tional park. Your target is to reach the village of Lahic settled by the Tat people coming originally from Iran. Mountains horses are the only way of transportation in the mountains, not only for people but also for different kind of material, wood, stones … . We will choose our route day by day in accordance with the weather, pathway and snow conditions. Each day will CAUCASUS EXPEDITIONS 2018 be an adventure and each trip will offer a new itinerary…be different and the choice is simply huge. Around our Caucasian cavalcades this trip offers a rich mix of nature and culture, dif- ferent kind of ecosystems and landscapes from the green mountains to the volcanic de- sert, old and new contrasting the country’s ancient heritage with its modern buildings in Baku, the pure unspoiled nature facing the former oil industrial lands. -

A New Record of the Persian Brook Salamander, Paradactylodon Persicus (Eiselt & Steiner, 1970) (Amphibia: Caudata: Hynobiidae) in Northern Iran
Bonn zoological Bulletin Volume 60 Issue 1 pp. 63–65 Bonn, May 2011 A new record of the Persian Brook Salamander, Paradactylodon persicus (Eiselt & Steiner, 1970) (Amphibia: Caudata: Hynobiidae) in northern Iran Faraham Ahmadzadeh1, 2*, Fatemeh Khanjani3, Aref Shadkam4 & Wolfgang Böhme2 1Department of Biodiversity and Ecosystem Management, Environmental Sciences Research Institute, Shahid Beheshti University, Evin, Tehran, G. C., Iran 2Herpetology Section, Zoologisches Forschungsmuseum Alexander Koenig (ZFMK), Adenauerallee 160, D-53113 Bonn, Germany 3Department of Habitat and Biodiversity, Faculty of Environment and Energy, Science and Research Branch, Islamic Azad University, Ponak, Tehran, Iran 4Department of Environment, Rezvan Shahr, Province Gilan, Iran *Corresponding Email address: [email protected]. INTRODUCTION The Persian Brook Salamander, Paradactylodon persicus 90 mm; tail length 120 mm; head large, 20 mm in length; (Eiselt & Steiner, 1970) is an endemic and poorly known vomerine teeth in two arch-shaped rows; snout rounded; species of northern Iran (Baloutch & Kami 1995; Kami fore and hind limbs with four digits; tail flattened later- 1999). It was originally described as Batrachuperus per- ally, with round-tapered end; dorsal head and body, as well sicus by Eiselt & Steiner (1970), but has been transferred as upper surface of tail brownish with yellow spots and to the genus Paradactylodon based on genetic studies by marblings; belly cream without pattern (Fig. 2a–b). Zhang et al. (2006). This species has been reported from two localities only: Weyser, southeast of Chalus, in Paradactylodon persicus inhabits the mountainous Mazandaran Province (36º 30´ 35” N and 51º 26´ 38” E) streams and brooks, with cool, fast-flowing water and Delmadeh village, southeast of Khalkhal, in Ardabil (Baloutch & Kami 1995; Kami 1999; Ahmadzadeh & Ka- Province (37º 22´ 34” N and 48º 47´ 35” E) (Kami 2004; mi 2009). -

Caudata: Hynobiidae): Heterochronies and Reductions
65 (1): 117 – 130 © Senckenberg Gesellschaft für Naturforschung, 2015. 4.5.2015 Development of the bony skeleton in the Taiwan salamander, Hynobius formosanus Maki, 1922 (Caudata: Hynobiidae): Heterochronies and reductions Anna B. Vassilieva 1 *, June-Shiang Lai 2, Shang-Fang Yang 2, Yu-Hao Chang 1 & Nikolay A. Poyarkov, Jr. 1 1 Department of Vertebrate Zoology, Biological Faculty, Lomonosov Moscow State University, Leninskiye Gory, GSP-1, Moscow 119991, Russia — 2 Department of Life Science, National Taiwan Normal University, 88, Sec. 4 Tingchou Rd., Taipei 11677, Taiwan, R.O.C. — *Cor- responding author; vassil.anna(at)gmail.com Accepted 19.ii.2015. Published online at www.senckenberg.de / vertebrate-zoology on 4.v.2015. Abstract The development of the bony skeleton in a partially embryonized lotic-breeding salamander Hynobius formosanus is studied using the ontogenetic series from late embryos to postmetamorphic juveniles and adult specimen. Early stages of skull development in this spe- cies are compared with the early cranial ontogeny in two non-embryonized lentic-breeding species H. lichenatus and H. nigrescens. The obtained results show that skeletal development distinguishes H. formosanus from other hynobiids by a set of important features: 1) the reduction of provisory ossifications (complete absence of palatine and reduced state of coronoid), 2) alteration of a typical sequence of ossification appearance, namely, the delayed formation of vomer and coronoid, and 3) the absence of a separate ossification center of a lacrimal and formation of a single prefrontolacrimal. These unique osteological characters in H. formosanus are admittedly connected with specific traits of its life history, including partial embryonization, endogenous feeding until the end of metamorphosis and relatively short larval period. -

Amphibiaweb's Illustrated Amphibians of the Earth
AmphibiaWeb's Illustrated Amphibians of the Earth Created and Illustrated by the 2020-2021 AmphibiaWeb URAP Team: Alice Drozd, Arjun Mehta, Ash Reining, Kira Wiesinger, and Ann T. Chang This introduction to amphibians was written by University of California, Berkeley AmphibiaWeb Undergraduate Research Apprentices for people who love amphibians. Thank you to the many AmphibiaWeb apprentices over the last 21 years for their efforts. Edited by members of the AmphibiaWeb Steering Committee CC BY-NC-SA 2 Dedicated in loving memory of David B. Wake Founding Director of AmphibiaWeb (8 June 1936 - 29 April 2021) Dave Wake was a dedicated amphibian biologist who mentored and educated countless people. With the launch of AmphibiaWeb in 2000, Dave sought to bring the conservation science and basic fact-based biology of all amphibians to a single place where everyone could access the information freely. Until his last day, David remained a tirelessly dedicated scientist and ally of the amphibians of the world. 3 Table of Contents What are Amphibians? Their Characteristics ...................................................................................... 7 Orders of Amphibians.................................................................................... 7 Where are Amphibians? Where are Amphibians? ............................................................................... 9 What are Bioregions? ..................................................................................10 Conservation of Amphibians Why Save Amphibians? ............................................................................. -
Comparative Osteology and Evolution of the Lungless Salamanders, Family Plethodontidae David B
COMPARATIVE OSTEOLOGY AND EVOLUTION OF THE LUNGLESS SALAMANDERS, FAMILY PLETHODONTIDAE DAVID B. WAKE1 ABSTRACT: Lungless salamanders of the family Plethodontidae comprise the largest and most diverse group of tailed amphibians. An evolutionary morphological approach has been employed to elucidate evolutionary rela tionships, patterns and trends within the family. Comparative osteology has been emphasized and skeletons of all twenty-three genera and three-fourths of the one hundred eighty-three species have been studied. A detailed osteological analysis includes consideration of the evolution of each element as well as the functional unit of which it is a part. Functional and developmental aspects are stressed. A new classification is suggested, based on osteological and other char acters. The subfamily Desmognathinae includes the genera Desmognathus, Leurognathus, and Phaeognathus. Members of the subfamily Plethodontinae are placed in three tribes. The tribe Hemidactyliini includes the genera Gyri nophilus, Pseudotriton, Stereochilus, Eurycea, Typhlomolge, and Hemidac tylium. The genera Plethodon, Aneides, and Ensatina comprise the tribe Pleth odontini. The highly diversified tribe Bolitoglossini includes three super genera. The supergenera Hydromantes and Batrachoseps include the nominal genera only. The supergenus Bolitoglossa includes Bolitoglossa, Oedipina, Pseudoeurycea, Chiropterotriton, Parvimolge, Lineatriton, and Thorius. Manculus is considered to be congeneric with Eurycea, and Magnadig ita is congeneric with Bolitoglossa. Two species are assigned to Typhlomolge, which is recognized as a genus distinct from Eurycea. No. new information is available concerning Haptoglossa. Recognition of a family Desmognathidae is rejected. All genera are defined and suprageneric groupings are defined and char acterized. Range maps are presented for all genera. Relationships of all genera are discussed. -

Spatial Diversity in the Diet of the Eurasian Eagle Owl Bubo Bubo in Iran
Podoces, 2014, 9(1): 7 –21 PODOCES 2014 Vol. 9, No. 1 Journal homepage: www.wesca.net Spatial Diversity in the Diet of the Eurasian Eagle Owl Bubo bubo in Iran Ján Obuch Comenius University in Bratislava, Detached Unit, SK-038 15 Blatnica, Slovakia. Article Info Abstract Original Research During five stays in Iran, the author collected remnants of the diet from seven species of owls. The most numerous were samples from the Eurasian Eagle Received 25 March 2014 Owl Bubo bubo , which were found in 38 sites, usually on rocky cliffs where Accepted 14 January 2015 the owls breed or where they roost during the day. A total of 7,862 items of prey were analysed. Mammals predominated (Mammalia, 56 species, Keywords 77.0%), and the species representation of birds was diverse (Aves, more than Eurasian Eagle Owl 100 species, 15.3%); lower vertebrates were hunted less often (Amphibia, Bubo bubo Reptilia, Pisces, 5.0%), while invertebrates (Evertebrata, 2.7%) were an Diet occasional food supplement. The most commonly represented rodents Iran (Rodentia) in the Elborz and Talysh Mountains were: Snow Vole Chionomys nivalis , Steppe Field Mouse Apodemus witherbyi and Common Vole Microtus obscurus; in the northern part of the Zagros Mountains: Brandt’s Hamster Mesocricetus brandti , Williams’ Jerboa Allactaga williamsi and Setzer’s Mouse-tailed Dormouse Myomimus setzeri ; in the central wetter part of the Zagros: Persian Jird Meriones persicus , Tristam’s Jird Meriones tristrami , Transcaucasian Mole Vole Ellobius lutescens and Grey Hamster Cricetulus migratorius ; in the drier part of the Zagros: Libyan Jird Meriones libycus , Sundevall’s Jird Meriones crassus and Indian Gerbil Tatera indica ; in the southern part of the Zagros in Fars Province: Iranian Vole Microtus irani , the rats Rattus rattus and R. -

Synchrotron Microtomography Applied to the Volumetric Analysis of Internal Structures of Thoropa Miliaris Tadpoles G
www.nature.com/scientificreports OPEN Synchrotron microtomography applied to the volumetric analysis of internal structures of Thoropa miliaris tadpoles G. Fidalgo1*, K. Paiva1, G. Mendes1, R. Barcellos1, G. Colaço2, G. Sena1, A. Pickler1, C. L. Mota1, G. Tromba3, L. P. Nogueira4, D. Braz5, H. R. Silva2, M. V. Colaço1 & R. C. Barroso1 Amphibians are models for studying applied ecological issues such as habitat loss, pollution, disease, and global climate change due to their sensitivity and vulnerability to changes in the environment. Developmental series of amphibians are informative about their biology, and X-ray based 3D reconstruction holds promise for quantifying morphological changes during growth—some with a direct impact on the possibility of an experimental investigation on several of the ecological topics listed above. However, 3D resolution and discrimination of their soft tissues have been difcult with traditional X-ray computed tomography, without time-consuming contrast staining. Tomographic data were initially performed (pre-processing and reconstruction) using the open- source software tool SYRMEP Tomo Project. Data processing and analysis of the reconstructed tomography volumes were conducted using the segmentation semi-automatic settings of the software Avizo Fire 8, which provide information about each investigated tissues, organs or bone elements. Hence, volumetric analyses were carried out to quantify the development of structures in diferent tadpole developmental stages. Our work shows that synchrotron X-ray microtomography using phase-contrast mode resolves the edges of the internal tissues (as well as overall tadpole morphology), facilitating the segmentation of the investigated tissues. Reconstruction algorithms and segmentation software played an important role in the qualitative and quantitative analysis of each target structure of the Thoropa miliaris tadpole at diferent stages of development, providing information on volume, shape and length. -

50 CFR Ch. I (10–1–20 Edition) § 16.14
§ 15.41 50 CFR Ch. I (10–1–20 Edition) Species Common name Serinus canaria ............................................................. Common Canary. 1 Note: Permits are still required for this species under part 17 of this chapter. (b) Non-captive-bred species. The list 16.14 Importation of live or dead amphib- in this paragraph includes species of ians or their eggs. non-captive-bred exotic birds and coun- 16.15 Importation of live reptiles or their tries for which importation into the eggs. United States is not prohibited by sec- Subpart C—Permits tion 15.11. The species are grouped tax- onomically by order, and may only be 16.22 Injurious wildlife permits. imported from the approved country, except as provided under a permit Subpart D—Additional Exemptions issued pursuant to subpart C of this 16.32 Importation by Federal agencies. part. 16.33 Importation of natural-history speci- [59 FR 62262, Dec. 2, 1994, as amended at 61 mens. FR 2093, Jan. 24, 1996; 82 FR 16540, Apr. 5, AUTHORITY: 18 U.S.C. 42. 2017] SOURCE: 39 FR 1169, Jan. 4, 1974, unless oth- erwise noted. Subpart E—Qualifying Facilities Breeding Exotic Birds in Captivity Subpart A—Introduction § 15.41 Criteria for including facilities as qualifying for imports. [Re- § 16.1 Purpose of regulations. served] The regulations contained in this part implement the Lacey Act (18 § 15.42 List of foreign qualifying breed- U.S.C. 42). ing facilities. [Reserved] § 16.2 Scope of regulations. Subpart F—List of Prohibited Spe- The provisions of this part are in ad- cies Not Listed in the Appen- dition to, and are not in lieu of, other dices to the Convention regulations of this subchapter B which may require a permit or prescribe addi- § 15.51 Criteria for including species tional restrictions or conditions for the and countries in the prohibited list. -

Quantifying Arabia–Eurasia Convergence Accommodated in the Greater Caucasus by Paleomagnetic Reconstruction ∗ A
Earth and Planetary Science Letters 482 (2018) 454–469 Contents lists available at ScienceDirect Earth and Planetary Science Letters www.elsevier.com/locate/epsl Quantifying Arabia–Eurasia convergence accommodated in the Greater Caucasus by paleomagnetic reconstruction ∗ A. van der Boon a, , D.J.J. van Hinsbergen a, M. Rezaeian b, D. Gürer a, M. Honarmand b, D. Pastor-Galán a,c, W. Krijgsman a, C.G. Langereis a a Utrecht University, The Netherlands b Institute for Advanced Studies in Basic Sciences, Zanjan, Iran c Center for Northeast Asian Studies, Tohoku University, Japan a r t i c l e i n f o a b s t r a c t Article history: Since the late Eocene, convergence and subsequent collision between Arabia and Eurasia was accommo- Received 23 June 2017 dated both in the overriding Eurasian plate forming the Greater Caucasus orogen and the Iranian plateau, Received in revised form 20 September and by subduction and accretion of the Neotethys and Arabian margin forming the East Anatolian plateau 2017 and the Zagros. To quantify how much Arabia–Eurasia convergence was accommodated in the Greater Accepted 9 November 2017 Caucasus region, we here provide new paleomagnetic results from 97 volcanic sites (∼500 samples) in Available online xxxx ∼ ◦ Editor: A. Yin the Talysh Mountains of NW Iran, that show 15 net clockwise rotation relative to Eurasia since the Eocene. We apply a first-order kinematic restoration of the northward convex orocline that formed to Keywords: the south of the Greater Caucasus, integrating our new data with previously published constraints on tectonic rotations rotations of the Eastern Pontides and Lesser Caucasus. -
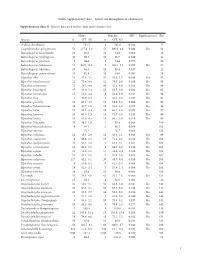
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius