Figure S1. Rice Genotype Selection Based on Shoot and Root Biomass
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
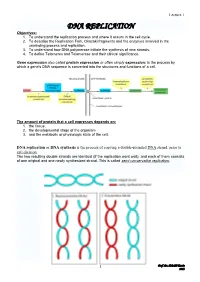
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

Light-Induced Psba Translation in Plants Is Triggered by Photosystem II Damage Via an Assembly-Linked Autoregulatory Circuit
Light-induced psbA translation in plants is triggered by photosystem II damage via an assembly-linked autoregulatory circuit Prakitchai Chotewutmontria and Alice Barkana,1 aInstitute of Molecular Biology, University of Oregon, Eugene, OR 97403 Edited by Krishna K. Niyogi, University of California, Berkeley, CA, and approved July 22, 2020 (received for review April 26, 2020) The D1 reaction center protein of photosystem II (PSII) is subject to mRNA to provide D1 for PSII repair remain obscure (13, 14). light-induced damage. Degradation of damaged D1 and its re- The consensus view in recent years has been that psbA transla- placement by nascent D1 are at the heart of a PSII repair cycle, tion for PSII repair is regulated at the elongation step (7, 15–17), without which photosynthesis is inhibited. In mature plant chloro- a view that arises primarily from experiments with the green alga plasts, light stimulates the recruitment of ribosomes specifically to Chlamydomonas reinhardtii (Chlamydomonas) (18). However, we psbA mRNA to provide nascent D1 for PSII repair and also triggers showed recently that regulated translation initiation makes a a global increase in translation elongation rate. The light-induced large contribution in plants (19). These experiments used ribo- signals that initiate these responses are unclear. We present action some profiling (ribo-seq) to monitor ribosome occupancy on spectrum and genetic data indicating that the light-induced re- cruitment of ribosomes to psbA mRNA is triggered by D1 photo- chloroplast open reading frames (ORFs) in maize and Arabi- damage, whereas the global stimulation of translation elongation dopsis upon shifting seedlings harboring mature chloroplasts is triggered by photosynthetic electron transport. -

A Web Tool for Hydrogenase Classification and Analysis Dan Søndergaard1, Christian N
www.nature.com/scientificreports OPEN HydDB: A web tool for hydrogenase classification and analysis Dan Søndergaard1, Christian N. S. Pedersen1 & Chris Greening2,3 H2 metabolism is proposed to be the most ancient and diverse mechanism of energy-conservation. The Received: 24 June 2016 metalloenzymes mediating this metabolism, hydrogenases, are encoded by over 60 microbial phyla Accepted: 09 September 2016 and are present in all major ecosystems. We developed a classification system and web tool, HydDB, Published: 27 September 2016 for the structural and functional analysis of these enzymes. We show that hydrogenase function can be predicted by primary sequence alone using an expanded classification scheme (comprising 29 [NiFe], 8 [FeFe], and 1 [Fe] hydrogenase classes) that defines 11 new classes with distinct biological functions. Using this scheme, we built a web tool that rapidly and reliably classifies hydrogenase primary sequences using a combination of k-nearest neighbors’ algorithms and CDD referencing. Demonstrating its capacity, the tool reliably predicted hydrogenase content and function in 12 newly-sequenced bacteria, archaea, and eukaryotes. HydDB provides the capacity to browse the amino acid sequences of 3248 annotated hydrogenase catalytic subunits and also contains a detailed repository of physiological, biochemical, and structural information about the 38 hydrogenase classes defined here. The database and classifier are freely and publicly available at http://services.birc.au.dk/hyddb/ Microorganisms conserve energy by metabolizing H2. Oxidation of this high-energy fuel yields electrons that can be used for respiration and carbon-fixation. This diffusible gas is also produced in diverse fermentation and 1 anaerobic respiratory processes . H2 metabolism contributes to the growth and survival of microorganisms across the three domains of life, including chemotrophs and phototrophs, lithotrophs and heterotrophs, aerobes and 1,2 anaerobes, mesophiles and extremophiles alike . -

Liver Med23 Ablation Improves Glucose and Lipid Metabolism Through Modulating FOXO1 Activity
Cell Research (2014) 24:1250-1265. npg © 2014 IBCB, SIBS, CAS All rights reserved 1001-0602/14 ORIGINAL ARTICLE www.nature.com/cr Liver Med23 ablation improves glucose and lipid metabolism through modulating FOXO1 activity Yajing Chu1, Leonardo Gómez Rosso1, Ping Huang2, Zhichao Wang1, Yichi Xu3, Xiao Yao1, Menghan Bao1, Jun Yan3, Haiyun Song2, Gang Wang1 1State Key Laboratory of Cell Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chi- nese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China; 2Key Laboratory of Nutrition and Metabolism, Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shang- hai 200031, China; 3CAS-MPG Partner Institute for Computational Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China Mediator complex is a molecular hub integrating signaling, transcription factors, and RNA polymerase II (RNAPII) machinery. Mediator MED23 is involved in adipogenesis and smooth muscle cell differentiation, suggesting its role in energy homeostasis. Here, through the generation and analysis of a liver-specific Med23-knockout mouse, we found that liver Med23 deletion improved glucose and lipid metabolism, as well as insulin responsiveness, and prevented diet-induced obesity. Remarkably, acute hepatic Med23 knockdown in db/db mice significantly improved the lipid profile and glucose tolerance. Mechanistically, MED23 participates in gluconeogenesis and cholesterol synthesis through modulating the transcriptional activity of FOXO1, a key metabolic transcription factor. Indeed, hepatic Med23 deletion impaired the Mediator and RNAPII recruitment and attenuated the expression of FOXO1 target genes. Moreover, this functional interaction between FOXO1 and MED23 is evolutionarily conserved, as the in vivo activities of dFOXO in larval fat body and in adult wing can be partially blocked by Med23 knockdown in Drosophila. -

Lecture Inhibition of Photosynthesis Inhibition at Photosystem I
1 Lecture Inhibition of Photosynthesis Inhibition at Photosystem I 1. General Information The popular misconception is that susceptible plants treated with these herbicides “starve to death” because they can no longer photosynthesize. In actuality, the plants die long before the food reserves are depleted. The photosynthetic inhibitors can be divided into two distinct groups, the inhibitors of Photosystem I and inhibitors of Photosystem II. Both of these groups work in the energy production step of photosynthesis, or the light reactions. Light is required as well as photosynthesis for these herbicides to kill susceptible plants. Herbicides that inhibit Photosystem I are considered to be contact herbicides and are often referred to as membrane disruptors. The end result is that cell membranes are rapidly destroyed resulting in leakage of cell contents into the intercellular spaces. These herbicides act as “electron interceptors” or “electron thieves” within Photosystem I of the light reaction of photosynthesis. They divert electrons from the normal electron transport sequence necessary in Photosystem I. This in turn inhibits photosynthesis. The membrane disruption occurs as a result of secondary responses. Herbicides that inhibit Photosystem I are represented by only one family, the bipyridyliums. See chemical structure shown under herbicide families. These molecules are cationic (positively charged) and are therefore highly water soluble. Their cationic properties also make them highly adsorbed to soil colloids resulting in no soil activity. 2. Mode of Action See Figure 7.1 (The electron transport chain in photosynthesis and the sites of action of herbicides that interfere with electron transfer in this chain (Q = electron acceptor; PQ = plastoquinone). -

Photosystem I-Based Applications for the Photo-Catalyzed Production of Hydrogen and Electricity
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 12-2014 Photosystem I-Based Applications for the Photo-catalyzed Production of Hydrogen and Electricity Rosemary Khuu Le University of Tennessee - Knoxville, [email protected] Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Biochemical and Biomolecular Engineering Commons Recommended Citation Le, Rosemary Khuu, "Photosystem I-Based Applications for the Photo-catalyzed Production of Hydrogen and Electricity. " PhD diss., University of Tennessee, 2014. https://trace.tennessee.edu/utk_graddiss/3146 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Rosemary Khuu Le entitled "Photosystem I- Based Applications for the Photo-catalyzed Production of Hydrogen and Electricity." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Doctor of Philosophy, with a major in Chemical Engineering. Paul D. Frymier, Major Professor We have read this dissertation and recommend its acceptance: Eric T. Boder, Barry D. Bruce, Hugh M. O'Neill Accepted for the Council: Carolyn R. Hodges Vice Provost and Dean of the Graduate School (Original signatures are on file with official studentecor r ds.) Photosystem I-Based Applications for the Photo-catalyzed Production of Hydrogen and Electricity A Dissertation Presented for the Doctor of Philosophy Degree The University of Tennessee, Knoxville Rosemary Khuu Le December 2014 Copyright © 2014 by Rosemary Khuu Le All rights reserved. -

(12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk Et Al
US 2014O155567A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk et al. (43) Pub. Date: Jun. 5, 2014 (54) MICROORGANISMS AND METHODS FOR (60) Provisional application No. 61/331,812, filed on May THE BIOSYNTHESIS OF BUTADENE 5, 2010. (71) Applicant: Genomatica, Inc., San Diego, CA (US) Publication Classification (72) Inventors: Mark J. Burk, San Diego, CA (US); (51) Int. Cl. Anthony P. Burgard, Bellefonte, PA CI2P 5/02 (2006.01) (US); Jun Sun, San Diego, CA (US); CSF 36/06 (2006.01) Robin E. Osterhout, San Diego, CA CD7C II/6 (2006.01) (US); Priti Pharkya, San Diego, CA (52) U.S. Cl. (US) CPC ................. CI2P5/026 (2013.01); C07C II/I6 (2013.01); C08F 136/06 (2013.01) (73) Assignee: Genomatica, Inc., San Diego, CA (US) USPC ... 526/335; 435/252.3:435/167; 435/254.2: (21) Appl. No.: 14/059,131 435/254.11: 435/252.33: 435/254.21:585/16 (22) Filed: Oct. 21, 2013 (57) ABSTRACT O O The invention provides non-naturally occurring microbial Related U.S. Application Data organisms having a butadiene pathway. The invention addi (63) Continuation of application No. 13/101,046, filed on tionally provides methods of using Such organisms to produce May 4, 2011, now Pat. No. 8,580,543. butadiene. Patent Application Publication Jun. 5, 2014 Sheet 1 of 4 US 2014/O155567 A1 ?ueudos!SMS |?un61– Patent Application Publication Jun. 5, 2014 Sheet 2 of 4 US 2014/O155567 A1 VOJ OO O Z?un61– Patent Application Publication US 2014/O155567 A1 {}}} Hººso Patent Application Publication Jun. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Sudips Revised Thesis
Investigation Of The Behavior Of The Gal4 Inhibitor Gal80 Of The GAL Genetic Switch In The Yeast Saccharomyces Cerevisiae Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Sudip Goswami, M.S. Graduate Program in Molecular Genetics The Ohio State University 2014 Dissertation Committee Dr. James Hopper, Advisor Dr. Stephen Osmani Dr. Hay-Oak Park Dr. Jian-Qiu Wu ii Copyright by Sudip Goswami 2014 iii ABSTRACT The DNA-binding transcriptional activator Gal4 and its regulators Gal80 and Gal3 constitute a galactose-responsive switch for the GAL genes of Saccharomyces cerevisiae. Gal4 binds to upstream activation sequences or UASGAL sites on GAL gene promoters as a dimer both in the absence and presence of galactose. In the absence of galactose, a Gal80 dimer binds to and masKs the Gal4 activation domain, inhibiting its activity. In the presence of galactose, Gal3 interacts with Gal80 and relieves Gal80’s inhibition of Gal4 activity allowing rapid induction of expression of GAL genes. In the first part of this work (Chapter 2) in-vitro chemical crosslinking coupled with SDS PAGE and native PAGE analysis were employed to show that the presence of Gal3 that can interact with Gal80 impairs Gal80 self association. In addition, live cell spinning disK confocal imaging showed that dissipation of newly discovered Gal80-2mYFP/2GFP clusters in galactose is dependent on Gal3’s ability to interact with Gal80. In the second part (Chapter 3), extensive analysis of Gal80 clusters was carried out which showed that these clusters associate strongly with the GAL1-10-7 locus and this association is dependent on the presence of the UASGAL sites at this locus. -

A Korarchaeal Genome Reveals Insights Into the Evolution of the Archaea
A korarchaeal genome reveals insights into the evolution of the Archaea James G. Elkinsa,b, Mircea Podarc, David E. Grahamd, Kira S. Makarovae, Yuri Wolfe, Lennart Randauf, Brian P. Hedlundg, Ce´ line Brochier-Armaneth, Victor Kunini, Iain Andersoni, Alla Lapidusi, Eugene Goltsmani, Kerrie Barryi, Eugene V. Koonine, Phil Hugenholtzi, Nikos Kyrpidesi, Gerhard Wannerj, Paul Richardsoni, Martin Kellerc, and Karl O. Stettera,k,l aLehrstuhl fu¨r Mikrobiologie und Archaeenzentrum, Universita¨t Regensburg, D-93053 Regensburg, Germany; cBiosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831; dDepartment of Chemistry and Biochemistry, University of Texas, Austin, TX 78712; eNational Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894; fDepartment of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520; gSchool of Life Sciences, University of Nevada, Las Vegas, NV 89154; hLaboratoire de Chimie Bacte´rienne, Unite´ Propre de Recherche 9043, Centre National de la Recherche Scientifique, Universite´de Provence Aix-Marseille I, 13331 Marseille Cedex 3, France; iU.S. Department of Energy Joint Genome Institute, Walnut Creek, CA 94598; jInstitute of Botany, Ludwig Maximilians University of Munich, D-80638 Munich, Germany; and kInstitute of Geophysics and Planetary Physics, University of California, Los Angeles, CA 90095 Communicated by Carl R. Woese, University of Illinois at Urbana–Champaign, Urbana, IL, April 2, 2008 (received for review January 7, 2008) The candidate division Korarchaeota comprises a group of uncul- and sediment samples from Obsidian Pool as an inoculum. The tivated microorganisms that, by their small subunit rRNA phylog- cultivation system supported the stable growth of a mixed commu- eny, may have diverged early from the major archaeal phyla nity of hyperthermophilic bacteria and archaea including an or- Crenarchaeota and Euryarchaeota. -

The Role of the Salvage Pathway in Nucleotide Sugar Biosynthesis
THE ROLE OF THE SALVAGE PATHWAY IN NUCLEOTIDE SUGAR BIOSYNTHESIS: IDENTIFICATION OF SUGAR KINASES AND NDP-SUGAR PYROPHOSPHORYLASES by TING YANG (Under the Direction of Maor Bar-Peled) ABSTRACT The synthesis of polysaccharides, glycoproteins, glycolipids, glycosylated secondary metabolites and hormones requires a large number of glycosyltransferases and a constant supply of nucleotide sugars. In plants, photosynthesis and the NDP-sugar inter-conversion pathway are the major entry points to form NDP-sugars. In addition to these pathways is the salvage pathway, a less understood metabolism that provides the flux of NDP-sugars. This latter pathway involves the hydrolysis of glycans to free sugars, sugar transport, sugar phosphorylation and nucleotidylation. The balance between glycan synthesis and recycling as well as its regulation at various plant developmental stages remains elusive as many of the molecular components are unknown. To understand how the salvage pathway contributes to the sugar flux and cell wall biosynthesis, my research focused on the functional identification of salvage pathway sugar kinases and NDP-sugar pyrophosphorylases. This research led to the first identification and enzymatic characterization of galacturonic acid kinase (GalA kinase), galactokinase (GalK), a broad UDP-sugar pyrophosphorylase (sloppy), two promiscuous UDP-GlcNAc pyrophosphorylases (GlcNAc-1-P uridylyltransferases), as well as UDP-sugar pyrophosphorylase paralogs from Trypanosoma cruzi and Leishmania major. To evaluate the salvage pathway in plant biology, we further investigated a sugar kinase mutant: galacturonic acid kinase mutant (galak) and determined if and how galak KO mutant affects the synthesis of glycans in Arabidopsis. Feeding galacturonic acid to the seedlings exhibited a 40-fold accumulation of free GalA in galak mutant, while the wild type (WT) plant readily metabolizes the fed-sugar.