Genetic Study of Scheduled Caste Populations of Tamil Nadu
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diverse Genetic Origin of Indian Muslims: Evidence from Autosomal STR Loci
Journal of Human Genetics (2009) 54, 340–348 & 2009 The Japan Society of Human Genetics All rights reserved 1434-5161/09 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Diverse genetic origin of Indian Muslims: evidence from autosomal STR loci Muthukrishnan Eaaswarkhanth1,2, Bhawna Dubey1, Poorlin Ramakodi Meganathan1, Zeinab Ravesh2, Faizan Ahmed Khan3, Lalji Singh2, Kumarasamy Thangaraj2 and Ikramul Haque1 The origin and relationships of Indian Muslims is still dubious and are not yet genetically well studied. In the light of historically attested movements into Indian subcontinent during the demic expansion of Islam, the present study aims to substantiate whether it had been accompanied by any gene flow or only a cultural transformation phenomenon. An array of 13 autosomal STR markers that are common in the worldwide data sets was used to explore the genetic diversity of Indian Muslims. The austere endogamy being practiced for several generations was confirmed by the genetic demarcation of each of the six Indian Muslim communities in the phylogenetic assessments for the markers examined. The analyses were further refined by comparison with geographically closest neighboring Hindu religious groups (including several caste and tribal populations) and the populations from Middle East, East Asia and Europe. We found that some of the Muslim populations displayed high level of regional genetic affinity rather than religious affinity. Interestingly, in Dawoodi Bohras (TN and GUJ) and Iranian Shia significant genetic contribution from West Asia, especially Iran (49, 47 and 46%, respectively) was observed. This divulges the existence of Middle Eastern genetic signatures in some of the contemporary Indian Muslim populations. -

Y-Chromosomal and Mitochondrial SNP Haplogroup Distribution In
Open Access Austin Journal of Forensic Science and Criminology Review Article Y-Chromosomal and Mitochondrial SNP Haplogroup Distribution in Indian Populations and its Significance in Disaster Victim Identification (DVI) - A Review Based Molecular Approach Sinha M1*, Rao IA1 and Mitra M2 1Department of Forensic Science, Guru Ghasidas Abstract University, India Disaster Victim Identification is an important aspect in mass disaster cases. 2School of Studies in Anthropology, Pt. Ravishankar In India, the scenario of disaster victim identification is very challenging unlike Shukla University, India any other developing countries due to lack of any organized government firm who *Corresponding author: Sinha M, Department of can make these challenging aspects an easier way to deal with. The objective Forensic Science, Guru Ghasidas University, India of this article is to bring spotlight on the potential and utility of uniparental DNA haplogroup databases in Disaster Victim Identification. Therefore, in this article Received: December 08, 2016; Accepted: January 19, we reviewed and presented the molecular studies on mitochondrial and Y- 2017; Published: January 24, 2017 chromosomal DNA haplogroup distribution in various ethnic populations from all over India that can be useful in framing a uniparental DNA haplogroup database on Indian population for Disaster Victim Identification (DVI). Keywords: Disaster Victim identification; Uniparental DNA; Haplogroup database; India Introduction with the necessity mentioned above which can reveal the fact that the human genome variation is not uniform. This inconsequential Disaster Victim Identification (DVI) is the recognized practice assertion put forward characteristics of a number of markers ranging whereby numerous individuals who have died as a result of a particular from its distribution in the genome, their power of discrimination event have their identity established through the use of scientifically and population restriction, to the sturdiness nature of markers to established procedures and methods [1]. -

Famine, Disease, Medicine and the State in Madras Presidency (1876-78)
FAMINE, DISEASE, MEDICINE AND THE STATE IN MADRAS PRESIDENCY (1876-78). LEELA SAMI UNIVERSITY COLLEGE LONDON DOCTOR OF PHILOSOPHY UMI Number: U5922B8 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. Dissertation Publishing UMI U592238 Published by ProQuest LLC 2013. Copyright in the Dissertation held by the Author. Microform Edition © ProQuest LLC. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code. ProQuest LLC 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106-1346 DECLARATION OF NUMBER OF WORDS FOR MPHIL AND PHD THESES This form should be signed by the candidate’s Supervisor and returned to the University with the theses. Name of Candidate: Leela Sami ThesisTitle: Famine, Disease, Medicine and the State in Madras Presidency (1876-78) College: Unversity College London I confirm that the following thesis does not exceed*: 100,000 words (PhD thesis) Approximate Word Length: 100,000 words Signed....... ... Date ° Candidate Signed .......... .Date. Supervisor The maximum length of a thesis shall be for an MPhil degree 60,000 and for a PhD degree 100,000 words inclusive of footnotes, tables and figures, but exclusive of bibliography and appendices. Please note that supporting data may be placed in an appendix but this data must not be essential to the argument of the thesis. -

Golden Research Thoughts
GRT Golden ReseaRch ThouGhTs ISSN: 2231-5063 Impact Factor : 4.6052 (UIF) Volume - 6 | Issue - 7 | January – 2017 ___________________________________________________________________________________ RAMANATHAPURAM : PAST AND PRESENT- A BIRD’S EYE VIEW Dr. A. Vadivel Department of History , Presidency College , Chennai , Tamil Nadu. ABSTRACT The present paper is an attempt to focus the physical features, present position and past history of the Ramnad District which was formed in the tail end of the Eighteenth Century. No doubt, the Ramnad District is the oldest district among the districts of the erstwhile Madras Presidency and the present Tamil Nadu. The District was formed by the British with the aim to suppress the southern poligars of the Tamil Country . For a while the southern poligars were rebellious against the expansion of the British hegemony in the south Tamil Country. After the formation of the Madras Presidency , this district became one of its districts. For sometimes it was merged with Madurai District and again its collectorate was formed in 1910. In the independent Tamil Nadu, it was trifurcated into Ramnad, Sivagangai and Viudhunagar districts. The district is, historically, a unique in many ways in the past and present. It was a war-torn region in the Eighteenth Century and famine affected region in the Nineteenth Century, and a region known for the rule of Setupathis. Many freedom fighters emerged in this district and contributed much for the growth of the spirit of nationalism. KEY WORDS : Ramanathapuram, Ramnad, District, Maravas, Setupathi, British Subsidiary System, Doctrine of Lapse ,Dalhousie, Poligars. INTRODUCTION :i Situated in the south east corner of Tamil Nadu State, Ramanathapuram District is highly drought prone and most backward in development. -

Community List
ANNEXURE - III LIST OF COMMUNITIES I. SCHEDULED TRIB ES II. SCHEDULED CASTES Code Code No. No. 1 Adiyan 2 Adi Dravida 2 Aranadan 3 Adi Karnataka 3 Eravallan 4 Ajila 4 Irular 6 Ayyanavar (in Kanyakumari District and 5 Kadar Shenkottah Taluk of Tirunelveli District) 6 Kammara (excluding Kanyakumari District and 7 Baira Shenkottah Taluk of Tirunelveli District) 8 Bakuda 7 Kanikaran, Kanikkar (in Kanyakumari District 9 Bandi and Shenkottah Taluk of Tirunelveli District) 10 Bellara 8 Kaniyan, Kanyan 11 Bharatar (in Kanyakumari District and Shenkottah 9 Kattunayakan Taluk of Tirunelveli District) 10 Kochu Velan 13 Chalavadi 11 Konda Kapus 14 Chamar, Muchi 12 Kondareddis 15 Chandala 13 Koraga 16 Cheruman 14 Kota (excluding Kanyakumari District and 17 Devendrakulathan Shenkottah Taluk of Tirunelveli District) 18 Dom, Dombara, Paidi, Pano 15 Kudiya, Melakudi 19 Domban 16 Kurichchan 20 Godagali 17 Kurumbas (in the Nilgiris District) 21 Godda 18 Kurumans 22 Gosangi 19 Maha Malasar 23 Holeya 20 Malai Arayan 24 Jaggali 21 Malai Pandaram 25 Jambuvulu 22 Malai Vedan 26 Kadaiyan 23 Malakkuravan 27 Kakkalan (in Kanyakumari District and Shenkottah 24 Malasar Taluk of Tirunelveli District) 25 Malayali (in Dharmapuri, North Arcot, 28 Kalladi Pudukkottai, Salem, South Arcot and 29 Kanakkan, Padanna (in the Nilgiris District) Tiruchirapalli Districts) 30 Karimpalan 26 Malayakandi 31 Kavara (in Kanyakumari District and Shenkottah 27 Mannan Taluk of Tirunelveli District) 28 Mudugar, Muduvan 32 Koliyan 29 Muthuvan 33 Koosa 30 Pallayan 34 Kootan, Koodan (in Kanyakumari District and 31 Palliyan Shenkottah Taluk of Tirunelveli District) 32 Palliyar 35 Kudumban 33 Paniyan 36 Kuravan, Sidhanar 34 Sholaga 39 Maila 35 Toda (excluding Kanyakumari District and 40 Mala Shenkottah Taluk of Tirunelveli District) 41 Mannan (in Kanyakumari District and Shenkottah 36 Uraly Taluk of Tirunelveli District) 42 Mavilan 43 Moger 44 Mundala 45 Nalakeyava Code III (A). -

Support Matarial Sociology Class-Xii 2016-17
SUPPORT MATARIAL SOCIOLOGY CLASS-XII 2016-17 Dr.Renu Bhatia S.K.V. Moti Bagh-I, New Delhi Principal and Group Leader Mr. A.M. Amir Ansari S.B.M. Sr. Sec. School (Govt. Sr. Lecture Sociology Aided) Shivaji Marg, N. Delhi-15 Ms. Sheema Banerjee Laxman Public School, Lecture Sociology Hauzkhas, New Delhi Mr. Zafar Hussain Anglo-Aravic, Sr. Sec. School Lecture Sociology Ajmeri Gate, Delhi Ms. Renu Shokeen Govt. Co-Ed., Sr. Sec. School PGT-Sociology Kanganheri, Delhi Ms. Anuradha Khanna Banyan Tree School, PGT-Sociology Lodhi Road, New Delhi-110003 Ms. Archana Arora SKV, A-Block, Jhangir Puri PGT-Sociology SOCIOLOGY (CODE NO.039) CLASS XII (2015-16) One Paper Theroy Marks 80 Unitwise Weightage 3 hours Units Periods Marks A Indian Society 1. Introducing Indian Society 10 32 2. Demographic Structure and Indain Society 10 Chapter-1 3. Social Institutions-Continuity and Change 12 and 4. Market as a Social Institution. 10 Chapter 7 5. Pattern of social Inequality and Exclusion 20 are non- 6. Challenges of Cultural Diversity 16 evaluative 7. Suggestions for Project Work 16 B Change and Developmentin Indian Society 8. Structural Change 10 9. Cultural Chage 12 10. The Story of Democaracy 18 Class XII - Sociology 2 11. Change and Development in Rural Society 10 48 12. Change and Development in industrial Society14 13. Globalization and Social Change 10 14. Mass Media and Communication 14 15. Social Movements 18 200 48 3 Class XII - Sociology BOOK I CHAPTER 2 THE DEMOGRAPCHIC STRUCTURE OF THE INDIAN SOCIETY KEY POINTS 1. Demography Demography, a systematic study of population, is a Greek term derived form two words, ‘demos’ (people) and graphein (describe) description of people. -

The Political Aco3mxddati0n of Primqpjdial Parties
THE POLITICAL ACO3MXDDATI0N OF PRIMQPJDIAL PARTIES DMK (India) and PAS (Malaysia) , by Y. Mansoor Marican M.Soc.Sci. (S'pore), 1971 A THESIS SUBMITTED IN PARTIAL FL^iDlMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES (Department of. Political Science) We accept this thesis as conforniing to the required standard THE IJNT^RSITY OF BRITISH COLUMBIA November. 1976 ® Y. Mansoor Marican, 1976. In presenting this thesis in partial fulfilment of the requirements for an advanced degree at the University of British Columbia, I agree that the Library shall make it freely available for reference and study. I further agree that permission for extensive copying of this thesis for scholarly purposes may be granted by the Head of my Department or by his representatives. It is understood that copying or publication of this thesis for financial gain shall not be allowed without my written permission. Department of POLITICAL SCIENCE The University of British Columbia 2075 Wesbrook Place Vancouver, Canada V6T 1W5 ABSTRACT This study is rooted in a theoretical interest in the development of parties that appeal mainly to primordial ties. The claims of social relationships based on tribe, race, language or religion have the capacity to rival the civil order of the state for the loyalty of its citizens, thus threatening to undermine its political authority. This phenomenon is endemic to most Asian and African states. Most previous research has argued that political competition in such contexts encourages the formation of primordially based parties whose activities threaten the integrity of these states. -
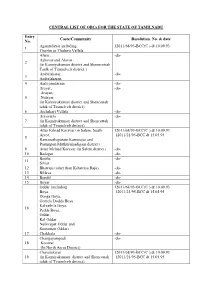
CENTRAL LIST of Obcs for the STATE of TAMILNADU Entry No
CENTRAL LIST OF OBC FOR THE STATE OF TAMILNADU E C/Cmm Rsoluti No. & da N. Agamudayar including 12011/68/93-BCC(C ) dt 10.09.93 1 Thozhu or Thuluva Vellala Alwar, -do- Azhavar and Alavar 2 (in Kanniyakumari district and Sheoncottah Taulk of Tirunelveli district ) Ambalakarar, -do- 3 Ambalakaran 4 Andi pandaram -do- Arayar, -do- Arayan, 5 Nulayar (in Kanniyakumari district and Shencottah taluk of Tirunelveli district) 6 Archakari Vellala -do- Aryavathi -do- 7 (in Kanniyakumari district and Shencottah taluk of Tirunelveli district) Attur Kilnad Koravar (in Salem, South 12011/68/93-BCC(C ) dt 10.09.93 Arcot, 12011/21/95-BCC dt 15.05.95 8 Ramanathapuram Kamarajar and Pasumpon Muthuramadigam district) 9 Attur Melnad Koravar (in Salem district) -do- 10 Badagar -do- Bestha -do- 11 Siviar 12 Bhatraju (other than Kshatriya Raju) -do- 13 Billava -do- 14 Bondil -do- 15 Boyar -do- Oddar (including 12011/68/93-BCC(C ) dt 10.09.93 Boya, 12011/21/95-BCC dt 15.05.95 Donga Boya, Gorrela Dodda Boya Kalvathila Boya, 16 Pedda Boya, Oddar, Kal Oddar Nellorepet Oddar and Sooramari Oddar) 17 Chakkala -do- Changayampadi -do- 18 Koravar (In North Arcot District) Chavalakarar 12011/68/93-BCC(C ) dt 10.09.93 19 (in Kanniyakumari district and Shencottah 12011/21/95-BCC dt 15.05.95 taluk of Tirunelveli district) Chettu or Chetty (including 12011/68/93-BCC(C ) dt 10.09.93 Kottar Chetty, 12011/21/95-BCC dt 15.05.95 Elur Chetty, Pathira Chetty 20 Valayal Chetty Pudukkadai Chetty) (in Kanniyakumari district and Shencottah taluk of Tirunelveli district) C.K. -

2021 Daily Prayer Guide for All People Groups & Unreached People
2021 Daily Prayer Guide for all People Groups & Unreached People Groups = LR-UPGs = of South Asia Joshua Project data, www.joshuaproject.net (India DPG is separate) AGWM Western edition I give credit & thanks to Create International for permission to use their PG photos. 2021 Daily Prayer Guide for all People Groups & LR-UPGs = Least-Reached-Unreached People Groups of South Asia = this DPG SOUTH ASIA SUMMARY: 873 total People Groups; 733 UPGs The 6 countries of South Asia (India; Bangladesh; Nepal; Sri Lanka; Bhutan; Maldives) has 3,178 UPGs = 42.89% of the world's total UPGs! We must pray and reach them! India: 2,717 total PG; 2,445 UPGs; (India is reported in separate Daily Prayer Guide) Bangladesh: 331 total PG; 299 UPGs; Nepal: 285 total PG; 275 UPG Sri Lanka: 174 total PG; 79 UPGs; Bhutan: 76 total PG; 73 UPGs; Maldives: 7 total PG; 7 UPGs. Downloaded from www.joshuaproject.net in September 2020 LR-UPG definition: 2% or less Evangelical & 5% or less Christian Frontier (FR) definition: 0% to 0.1% Christian Why pray--God loves lost: world UPGs = 7,407; Frontier = 5,042. Color code: green = begin new area; blue = begin new country "Prayer is not the only thing we can can do, but it is the most important thing we can do!" Luke 10:2, Jesus told them, "The harvest is plentiful, but the workers are few. Ask the Lord of the harvest, therefore, to send out workers into his harvest field." Why Should We Pray For Unreached People Groups? * Missions & salvation of all people is God's plan, God's will, God's heart, God's dream, Gen. -

The Historical Accomplishments of the Kallar Territory – a Study
Volume 1 Issue 2 February 2020 E-ISSN: 2582-2063 THE HISTORICAL ACCOMPLISHMENTS OF THE KALLAR TERRITORY – A STUDY Mr. A. MOORTHY RAJA Ph.D Research Scholar, Madurai Kamaraj University, Madurai, India Dr. C. CHELLAPPANDIAN Assistant Professor of History, Devanga Arts College, Aruppukottai, India Abstract In the present study, stating that the origin and historical background of kallars territory. Now-a-days, it is coined as Mukkulathor and that comprises the cluster of three identical clans of kallars, maravar and agamudaiyar. Here, the investigator has discussed to enhance the knowledge of kallars society and its features. They are numerically strong in the southern district of Tamilnadu. Keywords: Accomplishment, Mukkulathor, Territory, Ochchamma, Bejewelled, Goddess, Undeniably and Infanticide. Introduction The kallars can be said to be the longest living human tribe in India and one of the oldest in the world. They were most conservative section of the kallars and love of tradition. It is true in the sense that they had been in love with his tradition and had voluntarily anchored themselves to the ancients so that with them custom had been a deity to worship and conservatism had been their watchword. There is a very close connection among the three clans in their appearance, custom, manner, tradition and war like qualities. According to tradition, these people formed the important and strategic sections of the armies of the Tamil kings and Chief in olden days when fighting was even more an important profession then agriculture and supported a large population. But at present all the three have taken up farming. In spite of the alleged common ancestry, these three classes in early times formed themselves in to distinct castes and inter- marriage between the kallars and the other two th was not allowed. -

HIGH COURT of JUDICATURE at MADRAS FRIDAY 28 AUGUST 2020 INDEX Sl
. HIGH COURT OF JUDICATURE AT MADRAS FRIDAY 28 AUGUST 2020 INDEX Sl. Video Conference Sitting Arrangements / Coram Pages No. Court 1 NOTIFICATION NO. 183 / 2020 1 CHIEF JUSTICE & 2 VC 01 2 - 8 SENTHILKUMAR RAMAMOORTHY.J VINEET KOTHARI.J & 3 VC 02 9 - 10 KRISHNAN RAMASAMY.J R.SUBBIAH.J & 4 VC 03 11 - 16 C.SARAVANAN.J R.SUBBIAH.J & 5 VC 03 17 C.SARAVANAN.J N.KIRUBAKARAN.J & 6 VC 05 18 - 21 V.M.VELUMANI.J M.M.SUNDRESH.J & 7 VC 06 22 - 28 R.HEMALATHA.J T.S.SIVAGNANAM.J & 8 VC 07 29 - 31 PUSHPA SATHYANARAYANA.J 9 VC 08 M.DURAISWAMY.J 32 - 34 10 VC 09 T.RAJA.J 35 - 37 11 VC10 K.RAVICHANDRABAABU.J 38 - 42 12 VC 11 P.N.PRAKASH.J 43 - 45 13 VC 12 PUSHPA SATHYANARAYANA.J 46 - 47 14 VC 12 PUSHPA SATHYANARAYANA.J (Advance List) 48 - 57 15 VC 15 R.MAHADEVAN.J 58 - 70 16 VC 16 V.M.VELUMANI.J 71 - 72 17 VC 20 V.PARTHIBAN.J 73 - 81 18 VC 21 R.SUBRAMANIAN.J 82 - 85 19 VC 22 M.GOVINDARAJ.J 86 - 89 20 VC 23 M.SUNDAR.J (OS) 90 - 94 21 VC 26 M.S.RAMESH.J 95 - 103 22 VC 27 S.M.SUBRAMANIAM.J 104 - 107 23 VC 27 S.M.SUBRAMANIAM.J 108 - 110 24 VC 28 ANITA SUMANTH.J 111 - 120 25 VC 29 T.RAVINDRAN.J 121 - 122 26 VC 30 P.VELMURUGAN.J 123 - 126 27 VC 31 G.JAYACHANDRAN.J 127 - 129 28 VC 31 G.JAYACHANDRAN.J 130 - 133 29 VC 34 N.SATHISH KUMAR.J (OS) 134 - 142 30 VC 37 A.D.JAGADISH CHANDIRA.J 143 - 151 31 VC 39 ABDUL QUDDHOSE.J 152 - 157 32 VC 40 M.DHANDAPANI.J 158 - 161 33 VC 41 P.D.AUDIKESAVALU.J 162 - 165 34 VC 47 P.T.ASHA.J (OS) 166 - 174 35 VC 47 P.T.ASHA.J (Advance List for 31.08.2020) (OS) 175 - 179 36 VC 48 M.NIRMAL KUMAR.J 180 37 VC 48 M.NIRMAL KUMAR.J 181 - 184 38 VC 49 N.ANAND VENKATESH.J 185 - 193 39 VC 49 N.ANAND VENKATESH.J 194 - 195 40 VC 50 G.K.ILANTHIRAIYAN.J 196 - 200 41 VC 52 C.SARAVANAN.J 201 - 203 42 PROVISIONAL LIST 204 - 214 43 VC MC MASTER COURT 215 NOTIFICATION NO. -

Contribution of Pasumpon Muthuramalinga Thevar for the Welfare of De-Notified Tribes in Ramanathapuram District A-Study
Vol. 5 No. 4 April 2018 ISSN: 2321-788X UGC Approval No: 43960 Impact Factor: 3.025 CONTRIBUTION OF PASUMPON MUTHURAMALINGA THEVAR FOR THE WELFARE OF DE-NOTIFIED TRIBES IN RAMANATHAPURAM DISTRICT A-STUDY Article Particulars: Received: 15.03.2018 Accepted: 30.03.2018 Published: 28.04.2018 R. KUMARAVEL Ph.D. Research Scholar, Government Arts College (Autonomous), Karur, Tamil Nadu, India Abstract Pasumpon Muthuramalinga Thevar was known for his courage boldness and simplicity. Thevar fought for the cause of our National freedom and was an ardent follower of Nethaji subash Chandra Bose. Thevar’s words ignited the spirit of parriotism in the minds of the youth especially those in Madurai, Tirunelveli, and Ramanathapuram District. Thevar won the hearts of the youth by his eloquent and touching speeches. Thevar is well known as a patriot, a disciple of Nethaji and an undisputed leader of Forward Bloc and he is not only a leader of the Mukkilathors but also another communities. He was an unparallel leader in India, especially Ramamathapuram District in Tamil Nadu. He dedicated his entire life for upliftment of the down trodden people particularly depressed class people. He led the historic entry of Harijans into the Meenakshi Amman Temple In Madurai. He also worked for the upliftment of peasant class and fought to abolish the Zaminndar System. Keywords: Pasumpon Muthuramalinga Thevar, National freedom, Nethaji subash Chandra Bose, Ramanathapuram District, Forward Bloc, Harijans Meaning of De-Notified Tribes De-Notified Tribes (DNTs), also known as vimukta jati, are the tribes that were originally listed under the criminal Tribes Act of 1871, as Criminial Tribes and “addicted to the systematic commission of non-bail able offences’’.