Identification of a Nonsense Mutation in TNNI3K
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Co-Occupancy by Multiple Cardiac Transcription Factors Identifies
Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart Aibin Hea,b,1, Sek Won Konga,b,c,1, Qing Maa,b, and William T. Pua,b,2 aDepartment of Cardiology and cChildren’s Hospital Informatics Program, Children’s Hospital Boston, Boston, MA 02115; and bHarvard Stem Cell Institute, Harvard University, Cambridge, MA 02138 Edited by Eric N. Olson, University of Texas Southwestern, Dallas, TX, and approved February 23, 2011 (received for review November 12, 2010) Identification of genomic regions that control tissue-specific gene study of a handful of model genes (e.g., refs. 7–10), it has not been expression is currently problematic. ChIP and high-throughput se- evaluated using unbiased, genome-wide approaches. quencing (ChIP-seq) of enhancer-associated proteins such as p300 In this study, we used a modified ChIP-seq approach to define identifies some but not all enhancers active in a tissue. Here we genome wide the binding sites of these cardiac TFs (1). We show that co-occupancy of a chromatin region by multiple tran- provide unbiased support for collaborative TF interactions in scription factors (TFs) identifies a distinct set of enhancers. GATA- driving cardiac gene expression and use this principle to show that chromatin co-occupancy by multiple TFs identifies enhancers binding protein 4 (GATA4), NK2 transcription factor-related, lo- with cardiac activity in vivo. The majority of these multiple TF- cus 5 (NKX2-5), T-box 5 (TBX5), serum response factor (SRF), and “ binding loci (MTL) enhancers were distinct from p300-bound myocyte-enhancer factor 2A (MEF2A), here referred to as cardiac enhancers in location and functional properties. -

Comparison and Analysis on the Existing Single-Herbal Strategies Against Viral Myocarditis
Hindawi Genetics Research Volume 2021, Article ID 9952620, 12 pages https://doi.org/10.1155/2021/9952620 Research Article Comparison and Analysis on the Existing Single-Herbal Strategies against Viral Myocarditis Yu Cao ,1 Yang Liu,2 Tian Zhang,3 Jing Pan,4 Wei Lei,1 and Boli Zhang1 1Institute of Traditional Chinese Medicine, Tianjin University of Traditional Chinese Medicine, No. 10 Poyanghu Road, Tianjin 301617, China 2School of Chemical Engineering and Technology, Tianjin University, No. 135 Yaguan Road, Tianjin 300350, China 3State Key Laboratory of Dao-di Herbs, National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences, No. 16 Neinan Street, Beijing 100700, China 4Department of Reproductive Medicine, Inner Mongolia Maternal and Child Health Care Hospital, No. 18 North Second Ring Express Road, Hohhot 010020, China Correspondence should be addressed to Yu Cao; [email protected] Received 27 March 2021; Accepted 31 July 2021; Published 9 August 2021 Academic Editor: Hafiz Ishfaq Ahmad Copyright © 2021 Yu Cao et al. ,is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Purpose. Herbal medicine is one of crucial symbols of Chinese national medicine. Investigation on molecular responses of different herbal strategies against viral myocarditis is immeasurably conducive to targeting drug development in the current international absence of miracle treatment. Methods. Literature retrieval platforms were applied in the collection of existing empirical evidences for viral myocarditis-related single-herbal strategies. SwissTargetPrediction, Metascape, and Discovery Studio coordinating with multidatabases investigated underlying target genes, interactive proteins, and docking molecules in turn. -

TNNI3K Is a Novel Mediator of Myofilament Function and Phosphorylates Cardiac Troponin I
Brazilian Journal of Medical and Biological Research (2013) 46: 128-137, http://dx.doi.org/10.1590/1414-431X20122515 ISSN 1414-431X TNNI3K is a novel mediator of myofilament function and phosphorylates cardiac troponin I Hui Wang*, Lin Wang*, Li Song, Yan-Wan Zhang, Jue Ye, Rui-Xia Xu, Na Shi and Xian-Min Meng Core Laboratory, Fu Wai Hospital and Cardiovascular Institute, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China Abstract The phosphorylation of cardiac troponin I (cTnI) plays an important role in the contractile dysfunction associated with heart failure. Human cardiac troponin I-interacting kinase (TNNI3K) is a novel cardiac-specific functional kinase that can bind to cTnI in a yeast two-hybrid screen. The purpose of this study was to investigate whether TNNI3K can phosphorylate cTnI at specific sites and to examine whether the phosphorylation of cTnI caused by TNNI3K can regulate cardiac myofilament contractile function. Co-immunoprecipitation was performed to confirm that TNNI3K could interact with cTnI. Kinase assays further indicated that TNNI3K did not phosphorylate cTnI at Ser23/24 and Ser44, but directly phosphorylated Ser43 and Thr143 in vitro. The results obtained for adult rat cardiomyocytes also indicated that enhanced phosphorylation of cTnI at Ser43 and Thr143 correlated with rTNNI3K (rat TNNI3K) overexpression, and phosphorylation was reduced when rTNNI3K was knocked down. To determine the contractile function modulated by TNNI3K-mediated phosphorylation of cTnI, cardiomyocyte contraction was studied in adult rat ventricular myocytes. The contraction of cardiomyocytes increased with rTNNI3K overexpression and decreased with rTNNI3K knockdown. We conclude that TNNI3K may be a novel mediator of cTnI phosphorylation and contribute to the regulation of cardiac myofilament contraction function. -

Chromosome Abnormalities in Two Patients with Features of Autosomal Dominant Robinow Syndrome
ß 2007 Wiley-Liss, Inc. American Journal of Medical Genetics Part A 143A:1790–1795 (2007) Research Letter Chromosome Abnormalities in Two Patients With Features of Autosomal Dominant Robinow Syndrome Juliana F. Mazzeu,1 Ana Cristina Krepischi-Santos,1 Carla Rosenberg,1 Karoly Szuhai,2 Jeroen Knijnenburg,2 Janneke M.M. Weiss,3 Irina Kerkis,1 Zan Mustacchi,4 Guilherme Colin,5 Roˆmulo Mombach,6 Rita de Ca´ssia M. Pavanello,1 Paulo A. Otto,1 and Angela M. Vianna-Morgante1* 1Centro de Estudos do Genoma Humano, Departamento de Gene´tica e Biologia Evolutiva, Instituto de Biocieˆncias, Universidade de Sa˜o Paulo, Sa˜o Paulo, Brazil 2Department of Molecular Cell Biology, Leiden University Medical Center, Leiden, The Netherlands 3Department of Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands 4Hospital Infantil Darcy Vargas, Sa˜o Paulo, Brazil 5Departamento de Gene´tica Me´dica, Univille, Joinville, Brazil 6Centrinho Prefeito Luiz Gomes, Secretaria Municipal de Sau´de, Joinville, Brazil Received 13 April 2006; Accepted 13 December 2006 How to cite this article: Mazzeu JF, Krepischi-Santos AC, Rosenberg C, Szuhai K, Knijnenburg J, Weiss JMM, Kerkis I, Mustacchi Z, Colin G, Mombach R, Pavanello RM, Otto PA, Vianna-Morgante AM. 2007. Chromosome abnormalities in two patients with features of autosomal dominant Robinow syndrome. Am J Med Genet Part A 143A:1790–1795. To the Editor: Patient 1 Robinow syndrome [OMIM 180700] is characteriz- At age 3 4/12 years the girl was diagnosed as ed by fetal facies, mesomelic dwarfism, and hypo- affected by DRS (Fig. 1A). Detailed clinical examina- plastic genitalia. -

2021 Undergraduate Research Abstract Booklet
1 | P a g e Table of Contents FOREWORD ................................................................................................................................................... 4 Abidemi Awojuyigbe ..................................................................................................................................... 5 Aijalon Shantavia........................................................................................................................................... 6 Aminata Diagne ............................................................................................................................................. 8 The Exploration of BRAF Gene .................................................................................................................... 10 Araceli Estrada Martinez ............................................................................................................................. 10 Ashlee Young ............................................................................................................................................... 11 Ayanna D. Montegut ................................................................................................................................... 12 Brandon Bernäl ........................................................................................................................................... 13 Caleb Riggins .............................................................................................................................................. -

Page 1 Exploring the Understudied Human Kinome For
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.02.022277; this version posted June 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Exploring the understudied human kinome for research and therapeutic opportunities Nienke Moret1,2,*, Changchang Liu1,2,*, Benjamin M. Gyori2, John A. Bachman,2, Albert Steppi2, Rahil Taujale3, Liang-Chin Huang3, Clemens Hug2, Matt Berginski1,4,5, Shawn Gomez1,4,5, Natarajan Kannan,1,3 and Peter K. Sorger1,2,† *These authors contributed equally † Corresponding author 1The NIH Understudied Kinome Consortium 2Laboratory of Systems Pharmacology, Department of Systems Biology, Harvard Program in Therapeutic Science, Harvard Medical School, Boston, Massachusetts 02115, USA 3 Institute of Bioinformatics, University of Georgia, Athens, GA, 30602 USA 4 Department of Pharmacology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA 5 Joint Department of Biomedical Engineering at the University of North Carolina at Chapel Hill and North Carolina State University, Chapel Hill, NC 27599, USA Key Words: kinase, human kinome, kinase inhibitors, drug discovery, cancer, cheminformatics, † Peter Sorger Warren Alpert 432 200 Longwood Avenue Harvard Medical School, Boston MA 02115 [email protected] cc: [email protected] 617-432-6901 ORCID Numbers Peter K. Sorger 0000-0002-3364-1838 Nienke Moret 0000-0001-6038-6863 Changchang Liu 0000-0003-4594-4577 Ben Gyori 0000-0001-9439-5346 John Bachman 0000-0001-6095-2466 Albert Steppi 0000-0001-5871-6245 Page 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.04.02.022277; this version posted June 30, 2020. -

A Novel Missense Mutation in TNNI3K Causes Recessively Inherited Cardiac Conduction Disease in a Consanguineous Pakistani Family
G C A T T A C G G C A T genes Article A Novel Missense Mutation in TNNI3K Causes Recessively Inherited Cardiac Conduction Disease in a Consanguineous Pakistani Family Shafaq Ramzan 1,2, Stephanie Tennstedt 1,3,4, Muhammad Tariq 2 , Sheraz Khan 2 , Hafiza Noor Ul Ayan 1,2 , Aamir Ali 2 , Matthias Munz 1,3 , Holger Thiele 5, Asad Aslam Korejo 6, Abdul Razzaq Mughal 7, Syed Zahid Jamal 6, Peter Nürnberg 5,8 , Shahid Mahmood Baig 2,9,10, Jeanette Erdmann 1,3,4 and Ilyas Ahmad 1,3,4,* 1 Institute for Cardiogenetics, University of Lübeck, 23562 Lübeck, Germany; [email protected] (S.R.); [email protected] (S.T.); [email protected] (H.N.U.A.); [email protected] (M.M.); [email protected] (J.E.) 2 National Institute for Biotechnology and Genetic Engineering (NIBGE-C), Institute of Engineering and Applied Sciences (PIEAS), Islamabad 44000, Pakistan; [email protected] (M.T.); [email protected] (S.K.); [email protected] (A.A.); [email protected] (S.M.B.) 3 DZHK (German Research Centre for Cardiovascular Research) Partner Site Hamburg/Lübeck/Kiel, 23562 Lübeck, Germany 4 University Heart Center Lübeck, 23562 Lübeck, Germany 5 Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine, University Hospital Cologne, 50931 Cologne, Germany; [email protected] (H.T.); [email protected] (P.N.) 6 Citation: Ramzan, S.; Tennstedt, S.; National Institute of Cardiovascular Disease, Karachi 75510, Pakistan; [email protected] (A.A.K.); Tariq, M.; Khan, S.; Noor Ul Ayan, H.; [email protected] (S.Z.J.) 7 Faisalabad Institute of Cardiology, Faisalabad 38000, Pakistan; [email protected] Ali, A.; Munz, M.; Thiele, H.; Korejo, 8 Center for Molecular Medicine Cologne (CMMC), University of Cologne, Faculty of Medicine, A.A.; Mughal, A.R.; et al. -

Barkbase: Epigenomic Annotation of Canine Genomes
G C A T T A C G G C A T genes Article BarkBase: Epigenomic Annotation of Canine Genomes Kate Megquier 1, Diane P. Genereux 1 , Jessica Hekman 1 , Ross Swofford 1, Jason Turner-Maier 1, Jeremy Johnson 1, Jacob Alonso 1, Xue Li 1,2, Kathleen Morrill 1,2, Lynne J. Anguish 3, Michele Koltookian 1, Brittney Logan 2, Claire R. Sharp 4 , Lluis Ferrer 5, Kerstin Lindblad-Toh 1,6, Vicki N. Meyers-Wallen 7, Andrew Hoffman 8,9 and Elinor K. Karlsson 1,2,10,* 1 Vertebrate Genomics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; [email protected] (K.M.); [email protected] (D.P.G.); [email protected] (J.H.); swoff[email protected] (R.S.); [email protected] (J.T.-M.); [email protected] (J.J.); [email protected] (J.A.); [email protected] (X.L.); [email protected] (K.M.); [email protected] (M.K.); [email protected] (K.L.-T.) 2 Bioinformatics and Integrative Biology, University of Massachusetts Medical School, Worcester, MA 01655, USA; [email protected] 3 Baker Institute for Animal Health, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA; [email protected] 4 School of Veterinary and Life Sciences, College of Veterinary Medicine, Murdoch University, Perth, Murdoch, WA 6150, Australia; [email protected] 5 Departament de Medicina i Cirurgia Animals Veterinary School, Universitat Autonoma de Barcelona, 08193 Barcelona, Spain; [email protected] 6 Science for Life Laboratory, Department of Medical Biochemistry & -

Transcription of a Protein-Coding Gene on B Chromosomes of the Siberian
Trifonov et al. BMC Biology 2013, 11:90 http://www.biomedcentral.com/1741-7007/11/90 RESEARCH ARTICLE Open Access Transcription of a protein-coding gene on B chromosomes of the Siberian roe deer (Capreolus pygargus) Vladimir A Trifonov1*†, Polina V Dementyeva1†, Denis M Larkin2, Patricia CM O’Brien3, Polina L Perelman1, Fengtang Yang4, Malcolm A Ferguson-Smith3 and Alexander S Graphodatsky1 Abstract Background: Most eukaryotic species represent stable karyotypes with a particular diploid number. B chromosomes are additional to standard karyotypes and may vary in size, number and morphology even between cells of the same individual. For many years it was generally believed that B chromosomes found in some plant, animal and fungi species lacked active genes. Recently, molecular cytogenetic studies showed the presence of additional copies of protein-coding genes on B chromosomes. However, the transcriptional activity of these genes remained elusive. We studied karyotypes of the Siberian roe deer (Capreolus pygargus) that possess up to 14 B chromosomes to investigate the presence and expression of genes on supernumerary chromosomes. Results: Here, we describe a 2 Mbp region homologous to cattle chromosome 3 and containing TNNI3K (partial), FPGT, LRRIQ3 and a large gene-sparse segment on B chromosomes of the Siberian roe deer. The presence of the copy of the autosomal region was demonstrated by B-specific cDNA analysis, PCR assisted mapping, cattle bacterial artificial chromosome (BAC) clone localization and quantitative polymerase chain reaction (qPCR). By comparative analysis of B-specific and non-B chromosomal sequences we discovered some B chromosome-specific mutations in protein-coding genes, which further enabled the detection of a FPGT-TNNI3K transcript expressed from duplicated genes located on B chromosomes in roe deer fibroblasts. -

Role of TNNI3K in Cardiovascular Disease and Prospects for Therapy
UvA-DARE (Digital Academic Repository) Understanding cardiac electrical phenotypes in the genomic era Milano, A. Publication date 2015 Document Version Final published version Link to publication Citation for published version (APA): Milano, A. (2015). Understanding cardiac electrical phenotypes in the genomic era. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:27 Sep 2021 Chapter 4 TNNI3K in cardiovascular disease and prospects for therapy under revision at the Journal of Molecular and Cellular Cardiology Annalisa Milano, Elisabeth. M. Lodder, Connie. R. Bezzina 57 Chapter 4 Abstract Cardiovascular diseases are an important cause of morbidity and mortality worldwide and the global burden of these diseases continues to grow. Therefore new therapies are urgently needed. The role of protein kinases in disease, including cardiac disease, is long recognized, making kinases important therapeutic targets. -
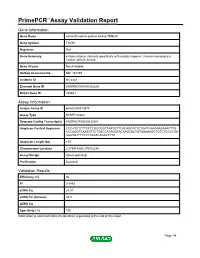
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name serine/threonine-protein kinase TNNI3K Gene Symbol Tnni3k Organism Rat Gene Summary kinase enzyme; interacts specifically with cardiac troponin I; human homolog is a cardiac specific kinase Gene Aliases Not Available RefSeq Accession No. NM_181769 UniGene ID Rn.3434 Ensembl Gene ID ENSRNOG00000028225 Entrez Gene ID 295531 Assay Information Unique Assay ID qRnoCID0013874 Assay Type SYBR® Green Detected Coding Transcript(s) ENSRNOT00000030341 Amplicon Context Sequence CCCATCTCTTCCTCGGTGGTTAATGTTCACAGCATTCTGATCAAGAAGAAATTTG ACCAGGTCAATGTTCTTGCCATAGGTACAAGCACTGTGAAAAGCTGTCTCGCTGA AGATGTTTTCCTTAGTCAGACTTTC Amplicon Length (bp) 105 Chromosome Location 2:279914520-279916240 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 96 R2 0.9993 cDNA Cq 24.51 cDNA Tm (Celsius) 80.5 gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Tnni3k, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Tissue-Based Alzheimer Gene Expression Markers–Comparison Of
Scheubert et al. BMC Bioinformatics 2012, 13:266 http://www.biomedcentral.com/1471-2105/13/266 RESEARCH ARTICLE Open Access Tissue-based Alzheimer gene expression markers–comparison of multiple machine learning approaches and investigation of redundancy in small biomarker sets Lena Scheubert1,3, Mitja Lustrekˇ 2, Rainer Schmidt3, Dirk Repsilber4* and Georg Fuellen3,5* Abstract Background: Alzheimer’s disease has been known for more than 100 years and the underlying molecular mechanisms are not yet completely understood. The identification of genes involved in the processes in Alzheimer affected brain is an important step towards such an understanding. Genes differentially expressed in diseased and healthy brains are promising candidates. Results: Based on microarray data we identify potential biomarkers as well as biomarker combinations using three feature selection methods: information gain, mean decrease accuracy of random forest and a wrapper of genetic algorithm and support vector machine (GA/SVM). Information gain and random forest are two commonly used methods. We compare their output to the results obtained from GA/SVM. GA/SVM is rarely used for the analysis of microarray data, but it is able to identify genes capable of classifying tissues into different classes at least as well as the two reference methods. Conclusion: Compared to the other methods, GA/SVM has the advantage of finding small, less redundant sets of genes that, in combination, show superior classification characteristics. The biological significance of the genes and gene pairs is discussed. Background could help both understanding the causes of the disease as Sporadic Alzheimer’s disease [1] is the most common well as suggest treatment options.