Molecular Data and Ploidal Levels Indicate Several
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Leaf Indumentum Types in Potentilla (Rosaceae) and Related Genera in Iran
Vol. 79, No. 2: 139-145, 2010 ACTA SOCIETATIS BOTANICORUM POLONIAE 139 LEAF INDUMENTUM TYPES IN POTENTILLA (ROSACEAE) AND RELATED GENERA IN IRAN MARZIEH BEYGOM FAGHIR1, FARIDEH ATTAR1, ALI FARAZMAND2, BARBARA ERTTER3, BENTE ERIKSEN4 1 Central Herbarium of Tehran University, School of Biology, University College of Science P.O. Box: 14155-6455, Tehran, Iran e-mail: [email protected] 2 University of Tehran, Department of Cell & Molecular Biology School of Biology, University College of Science P.O. Box: 14155-6455, Tehran, Iran 3 University and Jepson Herbaria, University of California, Berkeley California 94720-2465, USA 4 University of Göteborg, Department of Plant Environmental Sciences Box 461, SE-405 30, Göteborg Sweden (Received: December 5, 2009. Accepted: March 23, 2010) ABSTRACT Indumentum types of the leaves in 31 species of Potentilla L. (Rosaceae) and four related genera, especially Tylosperma Botsch., Schistophyllidium (Juz. ex Fed.) Ikonn., Drymocallis Fourr. ex Rydb., and Sibbaldia L. from Iran were investigated. Indumentum ultrastructure was studied by scanning electron microscopy (SEM). SEM observation revealed three type classes based on leaf indumentum: 1) straight (appressed erect); 2) straight-erect and crispate, and 3) crispate-floccose. The straight hair character (I type class) is widely distributed among all genera sampled and six sections of Potentilla. In contrast the crispate-floccose indumentum (III type class) is con- fined to all examined species of sections Speciosae and Pensylvanicae. While some sections especially Rectae (straight and straight crispate hairs) and Terminales (straight crispate and floccose crispate) posses two indu- mentum type classes. The present survey shows that indumentum types are of systematic importance and may form good key characters for identification purposes. -

Region 4 Threatened, Endangered and Sensitive Species List
INTERMOUNTAIN REGION (R4) THREATENED, ENDANGERED, PROPOSED, AND, SENSITIVE SPECIES June 2016 KNOWN / SUSPECTED DISTRIBUTION BY FOREST STATUS FOREST ENDANGERED ASH BOI B-T CAR CHA DIX FIS HUM M-L PAY SAL SAW TAR TOI UIN W-C MAMMALS Black-footed ferret 3/11/67 o o Mustela nigripes Sierra Nevada bighorn sheep Ovis canadensis X sierra January 3, 2000 BIRDS Southwestern willow flycatcher 2/27/95 X ? Empidonax traillii extimus ED 3/29/95 Whooping crane 3/11/67 X ? Grus americana REPTILES AND AMPHIBIANS Sierra Nevada Yellow-legged Frog 06/30/2014 X Rana sierrae INSECTS Mt. Charleston Blue Butterfly 10/21/2013 X Icaricia shasta charlestonensis FISH June sucker 3/31/86 o o Chasmistes liorus Bonytail chub 4/23/80 o o o o o o o Gila elegans Humpback chub 3/11/67 o o o o o o o Gila cypha Colorado pike minnow 3/11/67 o o o o o o o Ptychocheilus lucius Kendall Warm Springs dace 10/13/70 X Rhinichthys osculus Proposed, Endangered, Threatened, and Sensitive Species List, R4 Page 2 of 19 ENDANGERED ASH BOI B-T CAR CHA DIX FIS HUM M-L PAY SAL SAW TAR TOI UIN W-C Sockeye salmon, (Snake River0 11/20/91 + + + X Oncorhynchus nerka (CH 12/28/98) Razorback sucker 10/23/91 o o o o o o o Xyrauchen texanus (ED 11/22/91) Sturgeon, pallid o Scaphirhynchus albus PLANTS San Rafael cactus X Pediocactus despainii Clay phacelia 09/28/78 ? X Phacelia argillacea THREATENED ASH BOI B-T CAR CHA DIX FIS HUM M-L PAY SAL SAW TAR TOI UIN W-C MAMMALS Canada lynx 4/15/00 X X X X X X ? ? Lynx canadensis Grizzly bear 9/21/2009 X X Ursus arctos horribilis Gray wolf (Wyoming Rocky -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Botany Biological Evaluation
APPENDIX I Botany Biological Evaluation Biological Evaluation for Threatened, Endangered and Sensitive Plants and Fungi Page 1 of 35 for the Upper Truckee River Sunset Stables Restoration Project November 2009 UNITED STATES DEPARTMENT OF AGRICULTURE – FOREST SERVICE LAKE TAHOE BASIN MANAGEMENT UNIT Upper Truckee River Sunset Stables Restoration Project El Dorado County, CA Biological Evaluation for Threatened, Endangered and Sensitive Plants and Fungi PREPARED BY: ENTRIX, Inc. DATE: November 2009 APPROVED BY: DATE: _____________ Name, Forest Botanist, Lake Tahoe Basin Management Unit SUMMARY OF EFFECTS DETERMINATION AND MANAGEMENT RECOMMENDATIONS AND/OR REQUIREMENTS One population of a special-status bryophyte, three-ranked hump-moss (Meesia triquetra), was observed in the survey area during surveys on June 30, 2008 and August 28, 2008. The proposed action will not affect the moss because the population is located outside the project area where no action is planned. The following species of invasive or noxious weeds were identified during surveys of the Project area: cheatgrass (Bromus tectorum); bullthistle (Cirsium vulgare); Klamathweed (Hypericum perforatum); oxe-eye daisy (Leucanthemum vulgare); and common mullein (Verbascum Thapsus). The threat posed by these weed populations would not increase if the proposed action is implemented. An inventory and assessment of invasive and noxious weeds in the survey area is presented in the Noxious Weed Risk Assessment for the Upper Truckee River Sunset Stables Restoration Project (ENTRIX 2009). Based on the description of the proposed action and the evaluation contained herein, we have determined the following: There would be no significant effect to plant species listed as threatened, endangered, proposed for listing, or candidates under the Endangered Species Act of 1973, as amended (ESA), administered by the U.S. -

Hydrocotyle Sibthorpioides and H. Batrachium (Araliaceae) New for New York State
Atha, D. 2017. Hydrocotyle sibthorpioides and H. batrachium (Araliaceae) new for New York State. Phytoneuron 2017-56: 1– 6. Published 21 August 2017. ISSN 2153 733X HYDROCOTYLE SIBTHORPIOIDES AND H. BATRACHIUM (ARALIACEAE) NEW FOR NEW YORK STATE DANIEL ATHA Center for Conservation Strategy New York Botanical Garden Bronx, New York 10458 [email protected] ABSTRACT Spontaneous populations of Hydrocotyle sibthorpioides (lawn marsh pennywort) and H. batrachium (Araliaceae) are documented for New York state for the first time. Hydrocotyle batrachium is also new to North America. Hydrocotyle sibthorpioides was first found in 2013 in Queens county; H. batrachium was first found in 2016 in Westchester county. Both species are native to eastern Asia and show potential to be aggressive invaders in southeastern New York, particularly in wetlands. A key to the species of Hydrocotyle in New York State is provided. Prior to this report four species of Hydrocotyle were known from New York state, all of them native and all but one state-listed rarities: H. umbellata – Rare; H. verticillata var. verticillata – Endangered; and H. ranunculoides – Endangered (Young 2010). Among the native New York species, only H. americana occurs in abundance in the state. It is the only species historically reported for New York City and has not been documented for New York City since 1901. The present report documents two additional species for the New York flora, both native to Asia and naturalized in southeastern New York. Fertile herbarium specimens and DNA samples were obtained for all cited specimens and are available for analysis. Hydrocotyle sibthorpioides Lam. Spontaneous populations of Hydrocotyle sibthorpioides in New York were first detected and identified in Queens County by Nick Wagerik in the summer of 2013. -

Book of Abstracts.Pdf
1 List of presenters A A., Hudson 329 Anil Kumar, Nadesa 189 Panicker A., Kingman 329 Arnautova, Elena 150 Abeli, Thomas 168 Aronson, James 197, 326 Abu Taleb, Tariq 215 ARSLA N, Kadir 363 351Abunnasr, 288 Arvanitis, Pantelis 114 Yaser Agnello, Gaia 268 Aspetakis, Ioannis 114 Aguilar, Rudy 105 Astafieff, Katia 80, 207 Ait Babahmad, 351 Avancini, Ricardo 320 Rachid Al Issaey , 235 Awas, Tesfaye 354, 176 Ghudaina Albrecht , Matthew 326 Ay, Nurhan 78 Allan, Eric 222 Aydınkal, Rasim 31 Murat Allenstein, Pamela 38 Ayenew, Ashenafi 337 Amat De León 233 Azevedo, Carine 204 Arce, Elena An, Miao 286 B B., Von Arx 365 Bétrisey, Sébastien 113 Bang, Miin 160 Birkinshaw, Chris 326 Barblishvili, Tinatin 336 Bizard, Léa 168 Barham, Ellie 179 Bjureke, Kristina 186 Barker, Katharine 220 Blackmore, 325 Stephen Barreiro, Graciela 287 Blanchflower, Paul 94 Barreiro, Graciela 139 Boillat, Cyril 119, 279 Barteau, Benjamin 131 Bonnet, François 67 Bar-Yoseph, Adi 230 Boom, Brian 262, 141 Bauters, Kenneth 118 Boratyński, Adam 113 Bavcon, Jože 111, 110 Bouman, Roderick 15 Beck, Sarah 217 Bouteleau, Serge 287, 139 Beech, Emily 128 Bray, Laurent 350 Beech, Emily 135 Breman, Elinor 168, 170, 280 Bellefroid, Elke 166, 118, 165 Brockington, 342 Samuel Bellet Serrano, 233, 259 Brockington, 341 María Samuel Berg, Christian 168 Burkart, Michael 81 6th Global Botanic Gardens Congress, 26-30 June 2017, Geneva, Switzerland 2 C C., Sousa 329 Chen, Xiaoya 261 Cable, Stuart 312 Cheng, Hyo Cheng 160 Cabral-Oliveira, 204 Cho, YC 49 Joana Callicrate, Taylor 105 Choi, Go Eun 202 Calonje, Michael 105 Christe, Camille 113 Cao, Zhikun 270 Clark, John 105, 251 Carta, Angelino 170 Coddington, 220 Carta Jonathan Caruso, Emily 351 Cole, Chris 24 Casimiro, Pedro 244 Cook, Alexandra 212 Casino, Ana 276, 277, 318 Coombes, Allen 147 Castro, Sílvia 204 Corlett, Richard 86 Catoni, Rosangela 335 Corona Callejas , 274 Norma Edith Cavender, Nicole 84, 139 Correia, Filipe 204 Ceron Carpio , 274 Costa, João 244 Amparo B. -

Pala Park Habitat Assessment
Pala Park Bank Stabilization Project: Geotechnical Exploration TABLE OF CONTENTS SECTION 1.0 COUNTY OF RIVERSIDE ATTACHMENTS Biological Report Summary Report (Attachment E-3) Level of Significance Checklist (Attachment E-4) Biological Resources Map (Attachment E-5) Site Photographs (Attachment E-6) SECTION 2.0 HABITAT ASSESSMENT General Site Information ............................................................................................................... 1 Methods ........................................................................................................................................ 2 Existing Conditions ....................................................................................................................... 4 Special Status Resources ............................................................................................................. 8 Other Issues ................................................................................................................................ 14 Recommendations ...................................................................................................................... 14 References .................................................................................................................................. 16 LIST OF TABLES Page 1 Special Status Plant Species Known to Occur in the Vicinity of the Survey Area ........... 10 2 Chaparral Sand-Verbena Populations Observed in the Survey Area ............................. 12 3 Paniculate Tarplant -

San Bruno Mountain Habitat Conservation Plan Year
SAN BRUNO MOUNTAIN HABITAT CONSERVATION PLAN YEAR 2019-20 ACTIVITIES REPORT FOR FEDERALLY LISTED SPECIES Endangered Species 10(a)(1)(B) Permit TE215574-6 Prepared By: Evan Cole San Mateo County Parks Department 455 County Center, 4th Floor Redwood City, CA 94063 December 2020 TABLE OF CONTENTS Table of Contents I. INTRODUCTION ..............................................................................................................7 A. Covered Species Population Status ................................................................................... 7 2019 MISSION BLUE STATUS ......................................................................................................................... 8 2020 CALLIPPE SILVERSPOT STATUS.............................................................................................................. 8 2020 SAN BRUNO ELFIN STATUS ................................................................................................................... 9 RARE PLANT STATUS ..................................................................................................................................... 9 II. STATUS OF SPECIES OF CONCERN .........................................................................9 A. Mission Blue Butterfly (Icaricia icarioides missionensis) .............................................. 9 METHODOLOGY .......................................................................................................................................... 10 RESULTS ..................................................................................................................................................... -
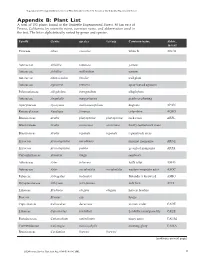
Vegetation and Ecological Characterisitics of Mixed-Conifer
Vegetation and Ecological Charactistics of Mixed-Conifer and Red Fir Forests at the Teakettle Experimental Forest Vegetation and Ecological Characteristics of Mixed-Conifer and Red Fir Forests at the Teakettle Experimental Forest Appendix B: Plant List A total of 152 plants found at the Teakettle Experimental Forest, 80 km east of Fresno, California, by scientific name, common name, and abbreviation used in the text. The list is alphabetically sorted by genus and species. Family Genus species var/ssp Common name Abbre. in text Pinaceae Abies concolor white fir ABCO Pinaceae Abies magnifica red fir ABMA Asteraceae Achillea lanulosa yarrow Asteraceae Achillea millefolium yarrow Asteraceae Adenocaulon bicolor trail plant Asteraceae Agroseris retrorsa spear-leaved agoseris Polemoniaceae Allophylum intregifolium allophylum Asteraceae Anaphalis margaritacea pearly everlasting Apocynaceae Apocynum androsaemifolium dogbane APAN Ranunculaceae Aquilegia formosa columbine AQFO Brassicaceae Arabis platysperma platysperma rock cress ARPL Brassicaceae Arabis rectissima rectissima bristly-leaved rock cress Brassicaceae Arabis repanda repanda repand rock cress Ericaceae Arctostaphylus nevadensis pinemat manzanita ARNE Ericaceae Arctostaphylus patula greenleaf manzanita ARPA Caryophyliaceae Arenaria kingii sandwort Asteraceae Aster foliaceus leafy aster ASFO Asteraceae Aster occidentalis occidentalis western mountain aster ASOC Fabaceae Astragalus bolanderi Bolander’s locoweed ASBO Dryopteridaceae Athryium felix-femina lady fern ATFI Liliaceae Brodiaea elegans elegans harvest brodeia Poaceae Bromus ssp. brome Cupressaceae Calocedrus decurrens incense cedar CADE Liliaceae Calochortus leichtlinii Leichtlin’s mariposa lily CALE Portulacaceae Calyptridium umbellatum pussy paws CAUM Convuvulaceae Calystegia malacophylla morning glory CAMA Brassicaceae Cardamine breweri breweri (continues on next page) 46 USDA Forest Service Gen.Tech. Rep. PSW-GTR-186. 2002. USDA Forest Service Gen.Tech. Rep. -

Lundberg Et Al. 2009
Molecular Phylogenetics and Evolution 51 (2009) 269–280 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Allopolyploidy in Fragariinae (Rosaceae): Comparing four DNA sequence regions, with comments on classification Magnus Lundberg a,*, Mats Töpel b, Bente Eriksen b, Johan A.A. Nylander a, Torsten Eriksson a,c a Department of Botany, Stockholm University, SE-10691, Stockholm, Sweden b Department of Environmental Sciences, Gothenburg University, Box 461, SE-40530, Göteborg, Sweden c Bergius Foundation, Royal Swedish Academy of Sciences, SE-10405, Stockholm, Sweden article info abstract Article history: Potential events of allopolyploidy may be indicated by incongruences between separate phylogenies Received 23 June 2008 based on plastid and nuclear gene sequences. We sequenced two plastid regions and two nuclear ribo- Revised 25 February 2009 somal regions for 34 ingroup taxa in Fragariinae (Rosaceae), and six outgroup taxa. We found five well Accepted 26 February 2009 supported incongruences that might indicate allopolyploidy events. The incongruences involved Aphanes Available online 5 March 2009 arvensis, Potentilla miyabei, Potentilla cuneata, Fragaria vesca/moschata, and the Drymocallis clade. We eval- uated the strength of conflict and conclude that allopolyploidy may be hypothesised in the four first Keywords: cases. Phylogenies were estimated using Bayesian inference and analyses were evaluated using conver- Allopolyploidy gence diagnostics. Taxonomic implications are discussed for genera such as Alchemilla, Sibbaldianthe, Cha- Fragariinae Incongruence maerhodos, Drymocallis and Fragaria, and for the monospecific Sibbaldiopsis and Potaninia that are nested Molecular phylogeny inside other genera. Two orphan Potentilla species, P. miyabei and P. cuneata are placed in Fragariinae. -

Chapter 6 ENUMERATION
Chapter 6 ENUMERATION . ENUMERATION The spermatophytic plants with their accepted names as per The Plant List [http://www.theplantlist.org/ ], through proper taxonomic treatments of recorded species and infra-specific taxa, collected from Gorumara National Park has been arranged in compliance with the presently accepted APG-III (Chase & Reveal, 2009) system of classification. Further, for better convenience the presentation of each species in the enumeration the genera and species under the families are arranged in alphabetical order. In case of Gymnosperms, four families with their genera and species also arranged in alphabetical order. The following sequence of enumeration is taken into consideration while enumerating each identified plants. (a) Accepted name, (b) Basionym if any, (c) Synonyms if any, (d) Homonym if any, (e) Vernacular name if any, (f) Description, (g) Flowering and fruiting periods, (h) Specimen cited, (i) Local distribution, and (j) General distribution. Each individual taxon is being treated here with the protologue at first along with the author citation and then referring the available important references for overall and/or adjacent floras and taxonomic treatments. Mentioned below is the list of important books, selected scientific journals, papers, newsletters and periodicals those have been referred during the citation of references. Chronicles of literature of reference: Names of the important books referred: Beng. Pl. : Bengal Plants En. Fl .Pl. Nepal : An Enumeration of the Flowering Plants of Nepal Fasc.Fl.India : Fascicles of Flora of India Fl.Brit.India : The Flora of British India Fl.Bhutan : Flora of Bhutan Fl.E.Him. : Flora of Eastern Himalaya Fl.India : Flora of India Fl Indi. -

A Natural Resource Condition Assessment for Sequoia and Kings Canyon National Parks Appendix 14 – Plants of Conservation Concern
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science A Natural Resource Condition Assessment for Sequoia and Kings Canyon National Parks Appendix 14 – Plants of Conservation Concern Natural Resource Report NPS/SEKI/ NRR—2013/665.14 In Memory of Rebecca Ciresa Wenk, Botaness ON THE COVER Giant Forest, Sequoia National Park Photography by: Brent Paull A Natural Resource Condition Assessment for Sequoia and Kings Canyon National Parks Appendix 14 – Plants of Conservation Concern Natural Resource Report NPS/SEKI/ NRR—2013/665.14 Ann Huber University of California Berkeley 41043 Grouse Drive Three Rivers, CA 93271 Adrian Das U.S. Geological Survey Western Ecological Research Center, Sequoia-Kings Canyon Field Station 47050 Generals Highway #4 Three Rivers, CA 93271 Rebecca Wenk University of California Berkeley 137 Mulford Hall Berkeley, CA 94720-3114 Sylvia Haultain Sequoia and Kings Canyon National Parks 47050 Generals Highway Three Rivers, CA 93271 June 2013 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate high-priority, current natural resource management information with managerial application. The series targets a general, diverse audience, and may contain NPS policy considerations or address sensitive issues of management applicability.