T Cell Development an Overview of the Intrathymic Intricacies Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Horse Gamma Globulin Stabilized in 0.3 Molar Glycine to a Ph of Approximately 6.8
LPD Reference: ATG-SIN-0414/1 Date of Last Revision: 08 September 2014 Country: Singapore Reference document: CDS Version 3.0, effective date: 17-Mar-2014 Reason for change: LPD update in accordance with CDS 3.0 ; 2014-09-08 IR: 1. Remove entire proposed subsection <Renal Transplant Prophylaxis> under ‘Pharmacodynamic Properties - Clinical Studies’ & 2. Relocate paragraph “Clinical trials […] standard supportive care alone” from <Therapeutic Indications> to < Pharmacodynamic Properties - Clinical Studies>, subsection <Aplastic Anemia>. Atgam® lymphocyte immune globulin, anti-thymocyte globulin [equine] sterile solution For Intravenous Use only DESCRIPTION ATGAM Sterile Solution contains lymphocyte immune globulin, anti-thymocyte globulin [equine]. It is the purified, concentrated, and sterile gamma globulin, primarily monomeric IgG, from hyperimmune serum of horses immunized with human thymus lymphocytes. Anti-thymocyte globulin (equine) is a transparent to slightly opalescent aqueous protein solution. It may appear colorless to faintly pink or brown and is nearly odorless. It may develop a slight granular or flaky deposit during storage. (For information about in-line filters, see Infusion Instructions in the POSOLOGY AND METHOD OF ADMINISTRATION.) Before release for clinical use, each lot of anti-thymocyte globulin (equine) is tested to assure its ability to inhibit rosette formation between human peripheral lymphocytes and sheep red blood cells in vitro. In each lot, antibody activity against human red blood cells and platelets is also measured and determined to be within acceptable limits. Only lots that test negative for antihuman serum protein antibody, antiglomerular basement membrane antibody and pyrogens are released. Each milliliter of anti-thymocyte globulin (equine) contains 50 mg of horse gamma globulin stabilized in 0.3 molar glycine to a pH of approximately 6.8. -

Road Safety Investment Program in Romania - AA-010269
CONSOLIDATED REPORT Framework agreement to support EIB advisory services (EIBAS) activities inside and outside EU-28 Lot 3: Transport Road Safety Investment Program in Romania - AA-010269 CONSULTING SAFEGE De Kleetlaan 5 B-1831 DIEGEM International Division SAFEGE SAS - SIÈGE SOCIAL Parc de l’Ile - 15/27 rue du Port 92022 NANTERRE CEDEX www.safege.com The authors take full responsibility for the contents of this report. The opinions expressed do not necessarily reflect the view of the Advisory Hub, nor the European Investment Bank, nor the European Commission Road Safety Investment Program in Romania - AA-010269 Framework agreement to support EIB advisory services (EIBAS) activities inside and outside EU-28 Lot 3: Transport Document quality information General information Author(s) Ilie Bricicaru, Kristiana Chakarova, Matthew Chamberlain, Loreta Robertina Gherman, Razvan Iulian Mazilu Project name RSIP in Romania – AA-010269 Document name Consolidated Report on Road Safety Investment Program in Romania Date April 05th, 2021 Reference Version 4 Addressee(s) Sent to: Name Organisation Sent on (date): Kevin CHEUNG, Per MATHIASEN, Teodora TATARU EIB Borislava GABROVSKA EIAH, EIB 05.04.2021 Cristian Andrei, Flavius PAVAL, Denyssa PPELIN CNAIR Copy to: Name Organisation Sent on (date): Joanna TALLEC, Spiros TRIANTAFILLOS SUEZ Consulting 05.04.2021 History of modifications Version Date Written by Approved & signed by: Ilie BRICICARU, Kristiana CHAKAROVA, Version 1 20.01.2021 Matthew CHAMBERLAIN Loreta Robertina GHERMAN Razvan Iulian MAZILU Ilie -

Our Immune System (Children's Book)
OurOur ImmuneImmune SystemSystem A story for children with primary immunodeficiency diseases Written by IMMUNE DEFICIENCY Sara LeBien FOUNDATION A note from the author The purpose of this book is to help young children who are immune deficient to better understand their immune system. What is a “B-cell,” a “T-cell,” an “immunoglobulin” or “IgG”? They hear doctors use these words, but what do they mean? With cheerful illustrations, Our Immune System explains how a normal immune system works and what treatments may be necessary when the system is deficient. In this second edition, a description of a new treatment has been included. I hope this book will enable these children and their families to explore together the immune system, and that it will help alleviate any confusion or fears they may have. Sara LeBien This book contains general medical information which cannot be applied safely to any individual case. Medical knowledge and practice can change rapidly. Therefore, this book should not be used as a substitute for professional medical advice. SECOND EDITION COPYRIGHT 1990, 2007 IMMUNE DEFICIENCY FOUNDATION Copyright 2007 by Immune Deficiency Foundation, USA. Readers may redistribute this article to other individuals for non-commercial use, provided that the text, html codes, and this notice remain intact and unaltered in any way. Our Immune System may not be resold, reprinted or redistributed for compensation of any kind without prior written permission from Immune Deficiency Foundation. If you have any questions about permission, please contact: Immune Deficiency Foundation, 40 West Chesapeake Avenue, Suite 308, Towson, MD 21204, USA; or by telephone at 1-800-296-4433. -

Il Calendario Dei Divieti Di Circolazione Della Grecia E Della Spagna Non È Ancora Disponibile
Driving restrictions, 2008 Austria 1. GENERAL DRIVING RESTRICTIONS Vehicles concerned Trucks with trailers, if the maximum authorised total weight of the motor vehicle or the trailer exceeds 3.5t; trucks, articulated vehicles and self-propelled industrial machines with an authorised total weight of more than 7.5t. Area Nationwide, with the exception of journeys made exclusively as part of a combined transport operation within a radius of 65km of the following transloading stations: Brennersee; Graz-Ostbahnhof; Salzburg-Hauptbahnhof; Wels-Verschiebebahnhof; Villach-Fürnitz; Wien-Südbahnhof; Wien-Nordwestbahnhof; Wörg; Hall in Tirol CCT; Bludenz CCT; Wolfurt CCT. Prohibition Saturdays from 15h00 to 24h00; Sundays and public holidays from 00h00 to 22h00 Public holidays 2008 1 January New Year’s Day 6 January Epiphany 24 March Easter Monday 1 May Labour Day; Ascension 12 May Whit Monday 22 May Corpus Christi 15 August Assumption 26 October National holiday 1 November All Saints’ Day 8 December Immaculate Conception 25 December Christmas Day 26 December Boxing Day Exceptions concerning trucks with trailers exceeding 3.5t · vehicles transporting milk; concerning vehicles with an authorised total weight of more than 7.5t · vehicles carrying meat or livestock for slaughter (but not the transport of heavy livestock on motorways), perishable foodstuffs (but not deep frozen goods), the supply of refreshments to tourist areas, urgent repairs to refrigeration plant, towing services (in all cases, according to § 46 StVO, it is obligatory to leave the motorway at the nearest exit), breakdown assistance vehicles, emergency vehicles, vehicles of a scheduled transport company (regular lines), and local trips on the two Saturdays preceding 24 December. -

1-Phosphate-Lyase-Deficient Mice Development in Sphingosine
Discontinued Postnatal Thymocyte Development in Sphingosine 1-Phosphate-Lyase-Deficient Mice This information is current as Claudia Weber, Andreas Krueger, Anika Münk, Constantin of October 1, 2021. Bode, Paul P. Van Veldhoven and Markus H. Gräler J Immunol 2009; 183:4292-4301; Prepublished online 11 September 2009; doi: 10.4049/jimmunol.0901724 http://www.jimmunol.org/content/183/7/4292 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2009/09/10/jimmunol.090172 Material 4.DC1 http://www.jimmunol.org/ References This article cites 48 articles, 20 of which you can access for free at: http://www.jimmunol.org/content/183/7/4292.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 1, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2009 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Discontinued Postnatal Thymocyte Development in Sphingosine 1-Phosphate-Lyase-Deficient Mice1,2 Claudia Weber,3* Andreas Krueger,3* Anika Mu¨nk,* Constantin Bode,* Paul P. -

Raport Preliminar Asupra Master Planului Pe Termen Scurt Mediu Si Lung
Raport preliminar asupra Master Planului pe termen scurt Mediu si Lung 19-08-2013 Preliminary Report on the Master Plan Short, Long and Medium Term 19-08- 2013 Transportation Guvernul României August 2013 Ministerul Transporturilor Master Planul General de Transport al României Versiune preliminară a Master Planului pe Termen Scurt, Mediu și Lung Elaborat de: ............................................................. Verificat de: Andrew Coates Simon Temple Frank Mohan Director Geoff Clarke Craig Bell Brian Vaughan Aprobat de: Martin Bright Director Error! Reference source not found.Raport Preliminar asupra Master Planului pe Termen Scurt, Mediu și Lung Rev No Comentarii Verificat de Aprobat de Data 1 Versiune preliminară SRT MJB 19/08/2013 Colmore Plaza, Colmore Circus Queensway, Birmingham, B4 6AT Telephone: 0121 262 1900 Website: http://www.aecom.com Job No: 60268467 Ref: Versiune preliminară Master Plan Data: August 2013 This document has been prepared by AECOM Limited for the sole use of our client (the “Client”) and in accordance with generally accepted consultancy principles, the budget for fees and the terms of reference agreed between AECOM Limited and the Client. Any information provided by third parties and referred to herein has not been checked or verified by AECOM Limited, unless otherwise expressly stated in the document. No third party may rely upon this document without the prior and express written agreement of AECOM Limited. 1 Cuprins 1 Scopul și obiectivele principale ale Master Planului .................................................................................................... -

Understanding the Immune System: How It Works
Understanding the Immune System How It Works U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES NATIONAL INSTITUTES OF HEALTH National Institute of Allergy and Infectious Diseases National Cancer Institute Understanding the Immune System How It Works U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES NATIONAL INSTITUTES OF HEALTH National Institute of Allergy and Infectious Diseases National Cancer Institute NIH Publication No. 03-5423 September 2003 www.niaid.nih.gov www.nci.nih.gov Contents 1 Introduction 2 Self and Nonself 3 The Structure of the Immune System 7 Immune Cells and Their Products 19 Mounting an Immune Response 24 Immunity: Natural and Acquired 28 Disorders of the Immune System 34 Immunology and Transplants 36 Immunity and Cancer 39 The Immune System and the Nervous System 40 Frontiers in Immunology 45 Summary 47 Glossary Introduction he immune system is a network of Tcells, tissues*, and organs that work together to defend the body against attacks by “foreign” invaders. These are primarily microbes (germs)—tiny, infection-causing Bacteria: organisms such as bacteria, viruses, streptococci parasites, and fungi. Because the human body provides an ideal environment for many microbes, they try to break in. It is the immune system’s job to keep them out or, failing that, to seek out and destroy them. Virus: When the immune system hits the wrong herpes virus target or is crippled, however, it can unleash a torrent of diseases, including allergy, arthritis, or AIDS. The immune system is amazingly complex. It can recognize and remember millions of Parasite: different enemies, and it can produce schistosome secretions and cells to match up with and wipe out each one of them. -
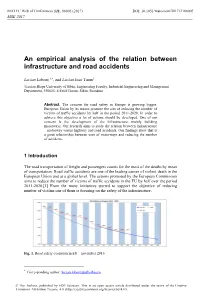
An Empirical Analysis of the Relation Between Infrastructure and Road Accidents
MATEC Web of Conferences 121, 06005 (2017) DOI: 10.1051/ matecconf/201712106005 MSE 2017 An empirical analysis of the relation between infrastructure and road accidents Lucian Lobonț 1,*, and Lucian Ioan Tarnu1 1Lucian Blaga University of Sibiu, Engineering Faculty, Industrial Engineering and Management Department, 550025, 4 Emil Cioran, Sibiu, România Abstract. The concern for road safety in Europe is growing bigger. European Union by its means promote the aim of reducing the number of victims of traffic accidents by half in the period 2011-2020. In order to achieve this objective a lot of actions should be developed. One of our concern is the development of the infrastructure, mainly building motorways. Our research aims to study the relation between infrastructure – motorway versus highway and road accidents. Our findings show that is a great relationship between uses of motorways and reducing the number of accidents. 1 Introduction The road transportation of freight and passengers counts for the most of the deaths by mean of transportation. Road traffic accidents are one of the leading causes of violent death in the European Union and at a global level. The actions promoted by the European Commission aims to reduce the number of victims of traffic accidents in the EU by half over the period 2011-2020.[1] From the many initiatives started to support the objective of reducing number of victims one of them is focusing on the safety of the infrastructure. Fig. 1. Road safety evolution in EU – november 2016 * Corresponding author: [email protected] © The Authors, published by EDP Sciences. This is an open access article distributed under the terms of the Creative Commons Attribution License 4.0 (http://creativecommons.org/licenses/by/4.0/). -

A Novel BCMA/CD3 Bispecific T-Cell Engager for the Treatment
OPEN Leukemia (2017) 31, 1743–1751 www.nature.com/leu ORIGINAL ARTICLE A novel BCMA/CD3 bispecific T-cell engager for the treatment of multiple myeloma induces selective lysis in vitro and in vivo S Hipp1, Y-T Tai2,3, D Blanset4, P Deegen5, J Wahl5, O Thomas5, B Rattel5, PJ Adam1, KC Anderson2,3 and M Friedrich5 B-cell maturation antigen (BCMA) is a highly plasma cell-selective protein that is expressed on malignant plasma cells of multiple myeloma (MM) patients and therefore is an ideal target for T-cell redirecting therapies. We developed a bispecific T-cell engager (BiTE) targeting BCMA and CD3ε (BI 836909) and studied its therapeutic impacts on MM. BI 836909 induced selective lysis of BCMA- positive MM cells, activation of T cells, release of cytokines and T-cell proliferation; whereas BCMA-negative cells were not affected. Activity of BI 836909 was not influenced by the presence of bone marrow stromal cells, soluble BCMA or a proliferation-inducing ligand (APRIL). In ex vivo assays, BI 836909 induced potent autologous MM cell lysis in both, newly diagnosed and relapsed/ refractory patient samples. In mouse xenograft studies, BI 836909 induced tumor cell depletion in a subcutaneous NCI-H929 xenograft model and prolonged survival in an orthotopic L-363 xenograft model. In a cynomolgus monkey study, administration of BI 836909 led to depletion of BCMA-positive plasma cells in the bone marrow. Taken together, these results show that BI 836909 is a highly potent and efficacious approach to selectively deplete BCMA-positive MM cells and represents a novel immunotherapeutic for the treatment of MM. -

REPORT Board of Directors 2019
REPORT Board of Directors 2019 1 Board of Directors Report for the year 2019 – Impact Group Content 03 2019 Highlights 04 Impact Group, overview 05 Projects portfolio 10 Activity of Impact Group in 2019 14 Residential real estate market 16 Strategy highlights 17 Financial results 19 Board of Directors and Executive Management 21 Main risks and uncertainties 22 Corporate Governance 24 Impact Developer & Contractor on the capital market 26 Reconciliation of IFRS Net Assets vs. EPRA 27 Annexes 27 Implementation of the Corporate Governance Code 34 Other information 35 Impact Group www.impactsa.ro 2 Board of Directors Report for the year 2019 – Impact Group 2019 HIGHLIGHTS Operational Financial IFRS NAV • 352 dwellings sold (30,185 sqm) • 263 pre-sale agreements and reservations on 31 Dec 584 mLEI 2019 estimated as sales in 2020 + 137.2 mLEI (2018: 446.8 mLEI) • Delivery of 192 apartments (14,500 sqm) for Greenfield Baneasa Residence (“Greenfield”) EPRA NAV • Further development of 500 apartments built in the Luxuria Domenii Residence project (“Luxuria”), to be delivered during 2020 839 mLEI • Starting works for the third phase of the Luxuria + 119 mLEI (2018: 720 mLEI) project, 130 apartments that will be delivered in the first half of 2021 Sales • Obtaining the new Zonal Urban Plan (PUZ) new developments in Greenfield Baneasa (Greenfield IV and Greenfield V), which includes Greenfield Plaza (a 157 mLEI commercial and leisure centre) + 56 mLEI (2018: 101 mLEI) • Obtaining the Zonal Urban Plan (PUZ) for the residential project Boreal Plus to -

PROHIBICIONES DE CIRCULACION PARA VEHICULOS PESADOS EN EUROPA ( Desde El 01.01.2011 Hasta El 31.12.2011 )
Madrid, 3 de Febrero 2011 ASTIC CIRCULAR Nº. 1.912 PROHIBICIONES DE CIRCULACION PARA VEHICULOS PESADOS EN EUROPA ( Desde el 01.01.2011 hasta el 31.12.2011 ) Ö EL INDICE de países figura en la última página (nº 58) A L E M A N I A Prohibición Para los vehículos de más de 7,5 t. de PMA, así como para camiones con remolque. Localización En toda la red de carreteras y autopistas. Período Domingos y días festivos de 00’00h. a 22’00h. Prohibición estival Localización Determinados tramos de autopistas y carreteras nacionales: - A1- de Cologne oeste vía Wuppertal, el cruce de Kamen y Münster hasta la salida a Cloppenburg, y de la entrada a Oyten hasta el cruce de Horst - A2- del cruce Oberhausen hasta el cruce Bad Oeynhausen - A3- del cruce Oberhausen hasta el cruce Cologne este, del cruce Mönchhof vía la intersección Francfort hasta la intersección Nürnberg - A4/E40- de la entrada a Herleshausen hasta el cruce Nossen - A5- del cruce Darmstadt vía Karlsruhe hasta el cruce Neuenburg - A6- de la entrada a Schwetzingen-Hockenheim hasta el cruce Nürnberg-sur - A7- de la entrada a Schleswig/Jagel hasta la salida Hambourg- Schnelsen-norte, del cruce Soltau-este al cruce Göttingen-nord; del cruce Schweinfurt/Werneck, el cruce Biebelried, el cruce Ulm/Elchingen, el cruce Allgäu hasta la frontera nacional en Füssen 2 - A8- del cruce Karlsruhe hasta la salida Munich-oeste y de la entrada a Munich- Ramersdorf hasta la salida Bad Reichenhall - A9/E51- circunvalación de Berlín (cruce Leipzig/cruce Potsdam) hasta la salida Munich-Schwabing - A10- circunvalación -

Colliers International Romania Mid-Year Market Update
H1 20 18 Colliers International Romania Mid-Year Market Update Accelerating success. 1 2 Colliers International Romania H1 Research & Forecast Report | 2018 Content Romania Macro Industrial Retail Update Market Market p. 04 p. 06 p. 08 Office Investment Land Market Market Market p. 10 p. 12 p. 16 Romania macro update In the post-crisis period, Romania has been the most we saw last year cannot be maintained and the anticipated The major challenges for the Romanian economy going successful economic convergence story in this part of slowdown is upon us. Some transitory factors weighed forward remain structural in nature (so more difficult Europe. In fact, if the service-led growth continues at a on GDP, leading to quasi-flat GDP readings in quarter-on- to tackle), like building highways, cutting back red tape pace similar to the post-crisis period, Romania is likely to quarter terms in 4Q17 and 1Q18, which is not something or corruption, increasing population activity rates and surpass Hungary by end-2022 and catch up to Slovakia we considered. The important note here is that due to improving education. Take the labour market for instance: by end-2028 in terms of GDP per capita, adjusted to the way economic growth is calculated and the statistical employers are finding it ever harder to fill in positions (both purchasing power standards (this indicator is widely used relevance of these quarters, it is looking nigh on impossible for white- and blue-collar positions), with unemployment as a proxy for living standards). to achieve an expansion rate above 5% in 2018 (barring near record lows of 4.4%.