S2 Table. List of Putative Candidate Genes Derived from the UMD 3.1 Assembly Involved In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Strategies to Increase ß-Cell Mass Expansion
This electronic thesis or dissertation has been downloaded from the King’s Research Portal at https://kclpure.kcl.ac.uk/portal/ Strategies to increase -cell mass expansion Drynda, Robert Lech Awarding institution: King's College London The copyright of this thesis rests with the author and no quotation from it or information derived from it may be published without proper acknowledgement. END USER LICENCE AGREEMENT Unless another licence is stated on the immediately following page this work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International licence. https://creativecommons.org/licenses/by-nc-nd/4.0/ You are free to copy, distribute and transmit the work Under the following conditions: Attribution: You must attribute the work in the manner specified by the author (but not in any way that suggests that they endorse you or your use of the work). Non Commercial: You may not use this work for commercial purposes. No Derivative Works - You may not alter, transform, or build upon this work. Any of these conditions can be waived if you receive permission from the author. Your fair dealings and other rights are in no way affected by the above. Take down policy If you believe that this document breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 Strategies to increase β-cell mass expansion A thesis submitted by Robert Drynda For the degree of Doctor of Philosophy from King’s College London Diabetes Research Group Division of Diabetes & Nutritional Sciences Faculty of Life Sciences & Medicine King’s College London 2017 Table of contents Table of contents ................................................................................................. -

Peptide, Peptidomimetic and Small Molecule Based Ligands Targeting Melanocortin Receptor System
PEPTIDE, PEPTIDOMIMETIC AND SMALL MOLECULE BASED LIGANDS TARGETING MELANOCORTIN RECEPTOR SYSTEM By ALEKSANDAR TODOROVIC A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2006 Copyright 2006 by Aleksandar Todorovic This document is dedicated to my family for everlasting support and selfless encouragement. ACKNOWLEDGMENTS I would like to thank and sincerely express my appreciation to all members, former and past, of Haskell-Luevano research group. First of all, I would like to express my greatest satisfaction by working with my mentor, Dr. Carrie Haskell-Luevano, whose guidance, expertise and dedication to research helped me reaching the point where I will continue the science path. Secondly, I would like to thank Dr. Ryan Holder who has taught me the principles of solid phase synthesis and initial strategies for the compounds design. I would like to thank Mr. Jim Rocca for the help and all necessary theoretical background required to perform proton 1-D NMR. In addition, I would like to thank Dr. Zalfa Abdel-Malek from the University of Cincinnati for the collaboration on the tyrosinase study project. Also, I would like to thank the American Heart Association for the Predoctoral fellowship that supported my research from 2004-2006. The special dedication and thankfulness go to my fellow graduate students within the lab and the department. iv TABLE OF CONTENTS page ACKNOWLEDGMENTS ................................................................................................ -

Analgesics and the Effects of Pharmacogenomics Disclosures: None
Cohen, Mindy, MD Analgesics and the Effects of Pharmacogenetics Analgesics and the Effects of Pharmacogenomics Disclosures: none Mindy Cohen, MD Learning Objectives 1. Review genetic variations that influence analgesic pharmacotherapy in children. 2. Identify the most common Before there was the need for polymorphisms in drug-metabolizing enzymes that influence analgesics. analgesia, there was… 3. Describe strategies for modifying analgesic regimens based on pharmacogenomics. PAIN Multifactorial Influences Genetic influence on Personality pain sensitivity Secondary Socio-economic gain status Pain Genetic influence on Genetics Environment analgesic medications Prior stress or trauma Cohen, Mindy, MD Analgesics and the Effects of Pharmacogenetics Genetic Influences on Pain Genetic Influences on Pain - Cases of Absent Pain - Twin Studies • Some rare cases explained by genetics • 2007- Thermal & chemical noxious stimuli • Loss-of-function mutations . 98 pairs of twins . α-subunit of voltage-gated sodium channel . 22-55% of variability was genetic . Other components that regulate functioning • 2008- Thermal noxious stimuli and homeostasis of nervous system . 96 twins . Cold-pressor pain • 7% of variability was genetic . Heat pain • 3% of variability was genetic Smith M et al. Clinical Genetics 2012 Norbury T et al. Brain 2007 Lotsch J et al. Trends in Pharm Sci 2010 Nielsen C et al. Pain 2008 Genetic Influences on Pain - Twin Studies • 2012- Thermal noxious stimuli, μ-agonists Analgesics and Genetics: . 112 pairs of twins . Pain tolerance and opioid analgesia Pharmacokinetics and . 24-60% of the response was influenced by Pharmacodynamics genetic makeup Angst M et al. Pain 2012 Genetic variation affects Genetic variation affects Pharmacokinetics Pharmacokinetics Cohen, Mindy, MD Analgesics and the Effects of Pharmacogenetics Pharmacokinetics Pharmacokinetics - Phase I Enzymes - Phase I Enzymes • Cytochrome P450 superfamily • Alter the chemical structure of drugs . -

Functionality and Genetics of Melanocortin and Purinergic Receptors
University of Latvia Faculty of Biology Vita Ignatoviča Doctoral Thesis Functionality and genetics of melanocortin and purinergic receptors Promotion to the degree of Doctor of Biology Molecular Biology Supervisor: Dr. Biol. Jānis Kloviņš Riga, 2012 1 The doctoral thesis was carried out in University of Latvia, Faculty of Biology, Department of Molecular biology and Latvian Biomedical Reseach and Study centre. From 2007 to 2012 The research was supported by Latvian Council of Science (LZPSP10.0010.10.04), Latvian Research Program (4VPP-2010-2/2.1) and ESF funding (1DP/1.1.1.2.0/09/APIA/VIAA/150 and 1DP/1.1.2.1.2/09/IPIA/VIAA/004). The thesis contains the introduction, 9 chapters, 38 subchapters and reference list. Form of the thesis: collection of articles in biology with subdiscipline in molecular biology Supervisor: Dr. biol. Jānis Kloviņš Reviewers: 1) Dr. biol., Prof. Astrīda Krūmiņa, Latvian Biomedical Reseach and Study centre 2) Dr. biol., Prof. Ruta Muceniece, University of Latvia, Department of Medicine, Pharmacy program 3) PhD Med, Assoc.Prof.David Gloriam, University of Copenhagen, Department of Drug Design and Pharmacology The thesis will be defended at the public section of the Doctoral Commitee of Biology, University of Latvia, in the conference hall of Latvian Biomedical Research and Study centre on July 6th, 2012, at 11.00. The thesis is available at the Library of the University of Latvia, Kalpaka blvd. 4. This thesis is accepted of the commencement of the degree of Doctor of Biology on April 19th, 2012, by the Doctoral Commitee of Biology, University of Latvia. -

Bitter Taste Perception in Neanderthals Through the Analysis of The
View metadata, citation and similar papers Downloadedat core.ac.uk from http://rsbl.royalsocietypublishing.org/ on March 22, 2016 brought to you by CORE provided by Repositorio Institucional de la Universidad de Oviedo Biol. Lett. (2009) 5, 809–811 The most extensively studied taste variation in doi:10.1098/rsbl.2009.0532 humans is sensitivity to a bitter substance called phe- Published online 12 August 2009 nylthiocarbamide (PTC). Although approximately 75 Evolutionary biology per cent of the world population perceives this sub- stance as intensely bitter, it is virtually tasteless for the remaining 25 per cent of the population (Kim & Bitter taste perception in Drayna 2004). This is owing to a dominant ‘taster’ allele that shows a similar frequency to the recessive Neanderthals through the ‘non-taster’ allele. PTC itself is not found in any vegetable, but chemically similar substances that analysis of the TAS2R38 produce an identical response to PTC are present in gene many plant foods (including Brussels sprouts, cabbage, broccoli and others). It was discovered Carles Lalueza-Fox1,*, Elena Gigli1, (Kim et al. 2003) that most of the variation in PTC Marco de la Rasilla2, Javier Fortea2 sensitivity is related to polymorphisms at the and Antonio Rosas3 TAS2R38 gene, a single 1002 bp coding exon that encodes a 333-amino-acid, G-protein-coupled recep- 1Institut de Biologia Evolutiva, CSIC-UPF, Dr. Aiguader 88, 08003 Barcelona, Spain tor. The TAS2R38 gene has three amino-acid changes 2A´ rea de Prehistoria, Departamento de Historia, Universidad de Oviedo, in high frequencies that determine only five main hap- Teniente Alfonso Martı´nez s/n, 33011 Oviedo, Spain lotypes. -

IL-34–Dependent Intrarenal and Systemic Mechanisms Promote Lupus Nephritis in MRL-Faslpr Mice
BASIC RESEARCH www.jasn.org IL-34–Dependent Intrarenal and Systemic Mechanisms Promote Lupus Nephritis in MRL-Faslpr Mice Yukihiro Wada,1 Hilda M. Gonzalez-Sanchez,1 Julia Weinmann-Menke,2 Yasunori Iwata,1 Amrendra K. Ajay,1 Myriam Meineck,2 and Vicki R. Kelley1 1Renal Division, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts; and 2Department of Nephrology and Rheumatology, University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany ABSTRACT lpr Background In people with SLE and in the MRL-Fas lupus mouse model, macrophages and autoanti- bodies are central to lupus nephritis. IL-34 mediates macrophage survival and proliferation, is expressed by tubular epithelial cells (TECs), and binds to the cFMS receptor on macrophages and to a newly identified second receptor, PTPRZ. Methods To investigate whether IL-34–dependent intrarenal and systemic mechanisms promote lupus lpr nephritis, we compared lupus nephritis and systemic illness in MRL-Fas mice expressing IL-34 and IL-34 lpr knockout (KO) MRL-Fas mice. We also assessed expression of IL-34 and the cFMS and PTPRZ receptors in patients with lupus nephritis. lpr Results Intrarenal IL-34 and its two receptors increase during lupus nephritis in MRL-Fas mice. In knock- out mice lacking IL-34, nephritis and systemic illness are suppressed. IL-34 fosters intrarenal macrophage accumulation via monocyte proliferation in bone marrow (which increases circulating monocytes that are recruited by chemokines into the kidney) and via intrarenal macrophage proliferation. This accumulation leads to macrophage-mediated TEC apoptosis. We also found suppression of circulating autoantibodies and glomerular antibody deposits in the knockout mice. -

Interleukin-34 Enhances the Tumor Promoting Function of Colorectal Cancer-Associated Fibroblasts
cancers Article Interleukin-34 Enhances the Tumor Promoting Function of Colorectal Cancer-Associated Fibroblasts Eleonora Franzè 1, Antonio Di Grazia 1, Giuseppe Sigismondo Sica 2, Livia Biancone 1, Federica Laudisi 1 and Giovanni Monteleone 1,* 1 Department of Systems Medicine, University of Rome “TOR VERGATA”, 00133 Rome, Italy; [email protected] (E.F.); [email protected] (A.D.G.); [email protected] (L.B.); [email protected] (F.L.) 2 Department of Surgery, University “TOR VERGATA” of Rome, 00133 Rome, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-06-7259-6158; Fax: +39-06-7259-6391 Received: 15 October 2020; Accepted: 24 November 2020; Published: 27 November 2020 Simple Summary: In colorectal cancer (CRC), cancer-associated fibroblasts (CAFs) promote tumor growth and progression through the synthesis of various molecules targeting the neoplastic cells. Here, we demonstrate that IL-34, a cytokine highly expressed in CRC tissue, regulates the function of CAFs in a paracrine and autocrine manner. Specifically, IL-34 induces normal fibroblasts (NFs) to acquire a cellular phenotype resembling that of CAFs, while IL-34 knockdown in CAFs reduces their tumorigenic properties and proliferation. Moreover, IL-34 stimulates NFs to produce netrin-1 and b-FGF—two factors that enhance CRC cell growth and migration. Altogether, our data support the involvement of IL-34 in CRC. Abstract: The stromal compartment of colorectal cancer (CRC) is marked by the presence of large numbers of fibroblasts, termed cancer-associated fibroblasts (CAFs), which promote CRC growth and progression through the synthesis of various molecules targeting the neoplastic cells. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Review of the Molecular Genetics of Basal Cell Carcinoma; Inherited Susceptibility, Somatic Mutations, and Targeted Therapeutics
cancers Review Review of the Molecular Genetics of Basal Cell Carcinoma; Inherited Susceptibility, Somatic Mutations, and Targeted Therapeutics James M. Kilgour , Justin L. Jia and Kavita Y. Sarin * Department of Dermatology, Stanford University School of Medcine, Stanford, CA 94305, USA; [email protected] (J.M.K.); [email protected] (J.L.J.) * Correspondence: [email protected] Simple Summary: Basal cell carcinoma is the most common human cancer worldwide. The molec- ular basis of BCC involves an interplay of inherited genetic susceptibility and somatic mutations, commonly induced by exposure to UV radiation. In this review, we outline the currently known germline and somatic mutations implicated in the pathogenesis of BCC with particular attention paid toward affected molecular pathways. We also discuss polymorphisms and associated phenotypic traits in addition to active areas of BCC research. We finally provide a brief overview of existing non-surgical treatments and emerging targeted therapeutics for BCC such as Hedgehog pathway inhibitors, immune modulators, and histone deacetylase inhibitors. Abstract: Basal cell carcinoma (BCC) is a significant public health concern, with more than 3 million cases occurring each year in the United States, and with an increasing incidence. The molecular basis of BCC is complex, involving an interplay of inherited genetic susceptibility, including single Citation: Kilgour, J.M.; Jia, J.L.; Sarin, nucleotide polymorphisms and genetic syndromes, and sporadic somatic mutations, often induced K.Y. Review of the Molecular Genetics of Basal Cell Carcinoma; by carcinogenic exposure to UV radiation. This review outlines the currently known germline and Inherited Susceptibility, Somatic somatic mutations implicated in the pathogenesis of BCC, including the key molecular pathways Mutations, and Targeted affected by these mutations, which drive oncogenesis. -
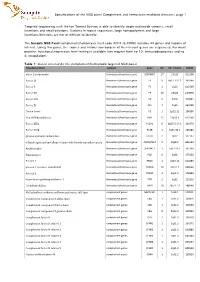
Specifications of the NGS Panel Complement and Hemostasis Mediated Diseases| Page 1
Specifications of the NGS panel Complement and hemostasis mediated diseases| page 1 Targeted sequencing with the Ion Torrent System is able to identify single nucleotide variants, small insertions and small deletions. Variants in repeat sequences, large homopolymers and large insertions/deletions are not or difficult to identify. The Sanquin NGS Panel complement/hemostasis (test code X001 to X006) includes 44 genes and regions of interest. Using this panel, the exones and intron/exon borders of the relevant genes are sequenced. For most proteins, functional/expression level testing is available (see request form no 10: immunodiagnostics and no 4: coagulation). Table 1: Genes covered by the complement/hemostasis targeted NGS panel. Encoded protein Context Gene Chr Chr. Positie OMIM alpha 2-antiplasmin Hemostasis/trombosis gene SERPINF2 17 17p13 613168 Factor IX Hemostasis/trombosis gene F9 X Xq27.1-27.2 300746 Factor V Hemostasis/trombosis gene F5 1 1q23 612309 Factor VII Hemostasis/trombosis gene F7 13 13q34 613878 Factor VIII Hemostasis/trombosis gene F8 X Xq28 300841 Factor XI Hemostasis/trombosis gene F11 4 4q35 264900 Tissue factor Hemostasis/trombosis gene F3 1 1p22-21 134390 Von Willebrand factor Hemostasis/trombosis gene VWF 12 12p13.3 613160 Factor XIIIa Hemostasis/trombosis gene F13A1 6 6p25.3-24.3 134570 Factor XIIIb Hemostasis/trombosis gene F13B 1 1q31-32.1 134580 gamma-glutamyl carboxylase Hemostasis/trombosis gene GGCX 2 2p12 137167 a Disintergrin and metalloproteinase wih thrombospondin repeats Hemostasis/trombosis gene -

Immunoregulatory Properties of the Cytokine IL-34
Cell. Mol. Life Sci. (2017) 74:2569–2586 DOI 10.1007/s00018-017-2482-4 Cellular and Molecular LifeSciences REVIEW Immunoregulatory properties of the cytokine IL-34 Carole Guillonneau1,2 · Séverine Bézie1,2 · Ignacio Anegon1,2 Received: 22 November 2016 / Revised: 10 January 2017 / Accepted: 30 January 2017 / Published online: 3 March 2017 © Springer International Publishing 2017 Abstract Interleukin-34 is a cytokine with only partially Introduction understood functions, described for the frst time in 2008. Although IL-34 shares very little homology with CSF-1 The CSF-1/CSF-1R interaction delivers a well-character- (CSF1, M-CSF), they share a common receptor CSF-1R ized signaling cascade leading in hematopoietic cells to (CSF-1R) and IL-34 has also two distinct receptors (PTP-ζ) proliferation, diferentiation, and function of the mono- and CD138 (syndecan-1). To make the situation more com- cytic lineage. The discovery in 2008 of IL-34, identifed by plex, IL-34 has also been shown as pairing with CSF-1 to screening of human protein library as a protein involved in form a heterodimer. Until now, studies have demonstrated monocyte viability [1] and subsequently, as a new ligand that this cytokine is released by some tissues that difer to of CSF-1R, has opened new perspectives. IL-34 actions those where CSF-1 is expressed and is involved in the dif- have been rendered more complex by the discovery of ferentiation and survival of macrophages, monocytes, and receptors for IL-34, others than CSF-1R: the receptor-type dendritic cells in response to infammation. -

Prion Disease and the Innate Immune System
Viruses 2012, 4, 3389-3419; doi:10.3390/v4123389 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Prion Disease and the Innate Immune System Barry M. Bradford and Neil A. Mabbott * The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Easter Bush Campus, Midlothian, EH25 9RG, UK; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +44-131-651-9100; Fax: +44-131-651-9105. Received: 6 October 2012; in revised form: 14 November 2012 / Accepted: 22 November 2012 / Published: 28 November 2012 Abstract: Prion diseases or transmissible spongiform encephalopathies are a unique category of infectious protein-misfolding neurodegenerative disorders. Hypothesized to be caused by misfolding of the cellular prion protein these disorders possess an infectious quality that thrives in immune-competent hosts. While much has been discovered about the routing and critical components involved in the peripheral pathogenesis of these agents there are still many aspects to be discovered. Research into this area has been extensive as it represents a major target for therapeutic intervention within this group of diseases. The main focus of pathological damage in these diseases occurs within the central nervous system. Cells of the innate immune system have been proven to be critical players in the initial pathogenesis of prion disease, and may have a role in the pathological progression of disease. Understanding how prions interact with the host innate immune system may provide us with natural pathways and mechanisms to combat these diseases prior to their neuroinvasive stage.