Single-Molecule Localization Algorithms in Super-Resolution Microscopy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Arrangement of a 4Pi Microscope for Reducing the Confocal Detection
Arrangement of a 4Pi microscope for reducing the confocal detection volume with two-photon excitation Nicolas Sandeau∗and Hugues Giovannini† Received 16 November 2005; received in revised form 7 February 2006; accepted 8 February 2006 Institut Fresnel, UMR 6133 CNRS, Universit´ePaul C´ezanne Aix-Marseille III, F-13397 Marseille cedex 20, France The main advantage of two-photon fluorescence confocal microscopy is the low absorption obtained with live tissues at the wavelengths of operation. However, the resolution of two-photon fluorescence confocal microscopes is lower than in the case of one-photon excitation. The 4Pi microscope type C working in two-photon regime, in which the excitation beams are coherently superimposed and, simultaneously, the emitted beams are also coherently added, has shown to be a good solution for increasing the resolution along the optical axis and for reducing the amplitude of the side lobes of the point spread function. However, the resolution in the transverse plane is poorer than in the case of one-photon excitation due to the larger wavelength in- volved in the two-photon fluorescence process. In this paper we show that a particular arrangement of the 4Pi microscope, referenced as 4Pi’ microscope, is a solution for obtaining a lateral resolution in the two-photon regime sim- ilar or even better to that obtained with 4Pi microscopes working in the arXiv:physics/0610036v1 [physics.bio-ph] 6 Oct 2006 one-photon excitation regime. Keywords: Resolution; Fluorescence microscopy; 4Pi microscopy; Confocal; Detection volume; Two-photon excitation ∗E-mail: [email protected] †E-mail: [email protected]; Tel: +33 491 28 80 66; Fax: +33 491 28 80 67 1 1 Introduction Strong efforts have been made in the last decade to improve the resolution of fluorescence microscopes. -

Cooperative 4Pi Excitation and Detection Yields Sevenfold Sharper Optical Sections in Live-Cell Microscopy
4146 Biophysical Journal Volume 87 December 2004 4146–4152 Cooperative 4Pi Excitation and Detection Yields Sevenfold Sharper Optical Sections in Live-Cell Microscopy Hilmar Gugel,* Jo¨rg Bewersdorf,y Stefan Jakobs,y Johann Engelhardt,z Rafael Storz,* and Stefan W. Hellyz *Leica Microsystems Heidelberg GmbH, 68165 Mannheim, Germany; yMax Planck Institute for Biophysical Chemistry, Department of NanoBiophotonics, 37070 Go¨ttingen, Germany; and zGerman Cancer Research Center, High Resolution Optical Microscopy Division, 69120 Heidelberg, Germany ABSTRACT Although the addition of just the excitation light field at the focus, or of just the fluorescence field at the detector is sufficient for a three- to fivefold resolution increase in 4Pi-fluorescence microscopy, substantial improvements of its optical properties are achieved by exploiting both effects simultaneously. They encompass not only an additional expansion of the optical bandwidth, but also an amplified transfer of the newly gained spatial frequencies to the image. Here we report on the realization and the imaging properties of this 4Pi microscopy mode of type C that also is the far-field microscope with the hitherto largest aperture. We show that in conjunction with two-photon excitation, the resulting optical transfer function displays a sevenfold improvement of axial three-dimensional resolution over confocal microscopy in aqueous samples, and more importantly, a marked transfer of all frequencies within its inner region of support. The latter is present also without the confocal pinhole. Thus, linear image deconvolution is possible both for confocalized and nonconfocalized live-cell 4Pi imaging. Realized in a state-of-the-art scanning microscope, this approach enables robust three-dimensional imaging of fixed and live cells at ;80 nm axial resolution. -

Dual-Color 4Pi-Confocal Microscopy with 3D-Resolution in the 100 Nm Range
Ultramicroscopy 90 (2002) 207–213 Dual-color 4Pi-confocal microscopy with 3D-resolution in the 100 nm range Hiroshi Kano, Stefan Jakobs, Matthias Nagorni, Stefan W. Hell* High Resolution Optical Microscopy Group, Max-Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37070 Gottin. gen, Germany Received 21 November 2000; received in revised form 2 July 2001 Abstract We report the development of simultaneous two-color channel recordingin 4Pi-confocal microscopy. A marked increase of spatial resolution over confocal microscopy becomes manifested in 4Pi-confocal three-dimensional (3D) data stacks of dual-labeled objects. The fundamentally improved resolution is verified both with densely labeled fluorescence beads as well as with membrane labeled fixed Escherichia coli. The synergistic combination of dual-color 4Pi-confocal recording with image restoration results in dual-color imaging with a 3D resolution in the 100 nm range. r 2002 Elsevier Science B.V. All rights reserved. 1. Introduction the typically 3.5 times sharper main maximum [1]. The side-lobes are removed mathematically by a By providinga fundamental improvement of linear filter, in which case a typical axial resolution axial sectioning, 4Pi-confocal microscopy [1] is a of 130–140 nm is achieved at a two-photon promisingdevelopment in three-dimensional (3D) excitation wavelength of 700–800 nm [3]. Alterna- far-field fluorescence microscopy. The 4Pi-confo- tively, the effective 3D-point spread function cal microscope owes its superior sectioningcap- (PSF) of the 4Pi-confocal microscope is used ability to the coherent addition of the spherical altogether to restore the 4Pi-confocal image data wavefronts produced by two opposingobjective with a non-linear restoration algorithm [4]. -

Two-Photon Excitation and Photoconversion of Eosfp in Dual-Color 4Pi Confocal Microscopy
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biophysical Journal Volume 92 June 2007 4451–4457 4451 Two-Photon Excitation and Photoconversion of EosFP in Dual-Color 4Pi Confocal Microscopy Sergey Ivanchenko,* Sylvia Glaschick,* Carlheinz Ro¨cker,* Franz Oswald,y Jo¨rg Wiedenmann,z and G. Ulrich Nienhaus*§ *Institute of Biophysics, yDepartment of Internal Medicine I, and zInstitute of General Zoology and Endocrinology, University of Ulm, 89069 Ulm, Germany; and §Department of Physics, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 ABSTRACT Recent years have witnessed enormous advances in fluorescence microscopy instrumentation and fluorescent marker development. 4Pi confocal microscopy with two-photon excitation features excellent optical sectioning in the axial di- rection, with a resolution in the 100 nm range. Here we apply this technique to cellular imaging with EosFP, a photoactivatable autofluorescent protein whose fluorescence emission wavelength can be switched from green (516 nm) to red (581 nm) by irradiation with 400-nm light. We have measured the two-photon excitation spectra and cross sections of the green and the red species as well as the spectral dependence of two-photon conversion. The data reveal that two-photon excitation and photo- activation of the green form of EosFP can be selectively performed by choosing the proper wavelengths. Optical highlighting of small subcellular compartments was shown on HeLa cells expressing EosFP fused to a mitochondrial targeting signal. After three-dimensionally confined two-photon conversion of EosFP within the mitochondrial networks of the cells, the converted re- gions could be resolved in a 3D reconstruction from a dual-color 4Pi image stack. -
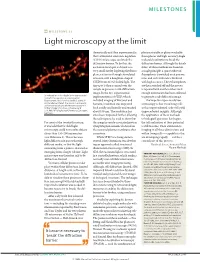
Milestone 21: Light Microscopy at the Limit
MILESTONES MILESTONE 21 Light microscopy at the limit theoretically and then experimentally, photoactivatable or photoswitchable that a stimulated-emission-depletion fluorophores and high-accuracy single- (STED) microscope can break the molecule localization to break the diffraction barrier. To do this, the diffraction barrier. Although the details excitation focal spot is shrunk to a differ, all three methods are based on very small size by depleting the fluoro- a single principle: a sparse subset of phores at its rim through stimulated fluorophores is switched on at any one emission with a doughnut-shaped time, and each molecule is localized STED beam of red-shifted light. The with high accuracy. These fluorophores tiny spot is then scanned over the are then switched off and the process sample to generate a sub-diffraction is repeated with another subset until image. In the first experimental enough information has been collected Stimulated-emission-depletion image recorded from a living neuron of an organotypical implementation of STED, which to generate a sub-diffraction image. hippocampal slice, showing dendritic spines in included imaging of live yeast and The hope for super-resolution unprecedented detail. The neuron is expressing yellow fluorescent protein. Image courtesy of K. bacteria, resolution was improved microscopy is that visualizing cells Willig, V. Nägerl, N. Urban, T. Bonhoeffer & both axially and laterally and reached at this unprecedented scale will yield S. W. Hell, MPI Biophysical Chemistry, Göttingen, Germany about 100 nm. The resolution has unprecedented insights. Although since been improved further, allowing the application of these methods the technique to be used to show that to biological questions has begun, For most of the twentieth century, the synaptic vesicle-associated protein the full realization of their potential it was held that far-field light synaptotagmin remains clustered on is still to come. -

Accurate 4Pi Single-Molecule Localization Using an Experimental PSF Model
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.18.997163; this version posted March 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Accurate 4Pi single-molecule localization using an experimental PSF model Yiming Li1,2, Elena Buglakova2,3, Yongdeng Zhang4, Jervis Vermal Thevathasan2,5, Joerg Bewersdorf4,6,7,8, Jonas Ries2,* 1Department of Biomedical Engineering, Southern University of Science and Technology, Shenzhen 518055, China 2European Molecular Biology Laboratory, Cell Biology and Biophysics, Heidelberg 69117, Germany 3Moscow Institute of Physics and Technology, Dolgoprudny 141701, Russia 4Department of Cell Biology, Yale School of Medicine, New Haven, CT 06511, USA 5Faculty of Biosciences, Heidelberg University, Heidelberg 69120, Germany 6Kavli Institute for Neuroscience, Yale School of Medicine, New Haven, CT 06511, USA 7Nanobiology Institute, Yale University, West Haven, CT 06516, USA 8Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA *Correspondence should be addressed to J.R. ([email protected]) Interferometric single-molecule localization microscopy (iPALM, 4Pi-SMS) uses multiphase interferometry to localize single fluorophores and achieves nanometer isotropic resolution in 3D. The current data analysis workflow, however, fails to reach the theoretical resolution limit due to the suboptimal localization algorithm. Here, we develop a method to fit an experimentally derived point spread function (PSF) model to the interference 4Pi-PSF. As the interference phase is not fixed with respect to the shape of the PSF, we decoupled the phase term in the model from the 3D position of the PSF. -

Assembly and Optimization of a Super-Resolution STORM Microscope for Nanoscopic Imaging of Biological Structures
Dissertation zur Erlangung des Doktorgrades der Fakult¨atf¨urChemie und Pharmazie der Ludwig-Maximilians-Universit¨atM¨unchen Assembly and optimization of a super-resolution STORM microscope for nanoscopic imaging of biological structures Jens Prescher aus M¨unchen, Deutschland 2016 Erkl¨arung Diese Dissertation wurde im Sinne von x7 der Promotionsordnung vom 28. November 2011 von Herrn Prof. Don C. Lamb, PhD betreut. Eidesstattliche Versicherung Diese Dissertation wurde eigenst¨andigund ohne unerlaubte Hilfe erarbeitet. M¨unchen, den 13. Januar 2016 ............................................ Dissertation eingereicht am 15. Januar 2016 1. Gutachter Prof. Don C. Lamb, PhD 2. Gutachter Prof. Dr. Christoph Br¨auchle M¨undliche Pr¨ufungam 18. Februar 2016 Contents Abstract v 1 Introduction 1 2 Theory of super-resolution fluorescence microscopy 5 2.1 Principles of fluorescence . 5 2.2 Fluorescence microscopy and optical resolution . 8 2.2.1 Principle of fluorescence microscopy . 9 2.2.2 Resolution in optical microscopy . 10 2.2.3 Total Internal Reflection Fluorescence Microscopy . 14 2.3 Super-resolution fluorescence microscopy . 17 2.3.1 Localization-based super-resolution microscopy . 18 2.3.1.1 Photoactivated Localization Microscopy (PALM) . 21 2.3.1.2 Stochastic Optical Reconstruction Microscopy (STORM) . 22 2.3.1.3 Direct Stochastic Optical Reconstruction Microscopy (dSTORM) 23 2.3.2 Determination of resolution in STORM and PALM . 24 2.3.3 Three-dimensional localization-based super-resolution imaging . 25 2.3.4 Overview over other optical super-resolution methods . 27 3 Experimental setup and analysis methods for STORM 29 3.1 Experimental setup . 29 3.1.1 Instrumentation of the STORM setup . -

Can Developments in Tissue Optical Clearing Aid Super-Resolution Microscopy Imaging?
International Journal of Molecular Sciences Review Can Developments in Tissue Optical Clearing Aid Super-Resolution Microscopy Imaging? Paweł Matryba 1,2,3,*, Kacper Łukasiewicz 4 , Monika Pawłowska 3,5 , Jacek Tomczuk 1 and Jakub Goł ˛ab 1 1 Department of Immunology, Medical University of Warsaw, 02-097 Warsaw, Poland; [email protected] (J.T.); [email protected] (J.G.) 2 The Doctoral School of the Medical University of Warsaw, Medical University of Warsaw, 02-097 Warsaw, Poland 3 Laboratory of Neurobiology, BRAINCITY, Nencki Institute of Experimental Biology of Polish Academy of Sciences, 02-093 Warsaw, Poland; [email protected] 4 Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA 95064, USA; [email protected] 5 Institute of Experimental Physics, Faculty of Physics, University of Warsaw, 02-093 Warsaw, Poland * Correspondence: [email protected] Abstract: The rapid development of super-resolution microscopy (SRM) techniques opens new avenues to examine cell and tissue details at a nanometer scale. Due to compatibility with specific labelling approaches, in vivo imaging and the relative ease of sample preparation, SRM appears to be a valuable alternative to laborious electron microscopy techniques. SRM, however, is not free from drawbacks, with the rapid quenching of the fluorescence signal, sensitivity to spherical aberrations and light scattering that typically limits imaging depth up to few micrometers being the most pronounced ones. Recently presented and robustly optimized sets of tissue optical clearing (TOC) techniques turn biological specimens transparent, which greatly increases the tissue thickness Citation: Matryba, P.; Łukasiewicz, K.; Pawłowska, M.; Tomczuk, J.; that is available for imaging without loss of resolution. -

High Resolution Imaging of Live Mitochondria ⁎ Stefan Jakobs
Biochimica et Biophysica Acta 1763 (2006) 561–575 http://www.elsevier.com/locate/bba Review High resolution imaging of live mitochondria ⁎ Stefan Jakobs Max-Planck-Institute for Biophysical Chemistry, Department of NanoBiophotonics, Mitochondrial Structure and Dynamics Group, Am Fassberg 11, 37077 Goettingen, Germany Received 16 January 2006; received in revised form 12 April 2006; accepted 13 April 2006 Available online 20 April 2006 Abstract Classically, mitochondria have been studied by biochemical, genetic and electron microscopic approaches. In the last two decades, it became evident that mitochondria are highly dynamic organelles that are frequently dividing and fusing, changing size and shape and traveling long distances throughout the life of a cell. The study of the complex structural changes of mitochondria in vivo became possible with the advent of fluorescent labeling techniques in combination with live cell imaging microscopy. This review aims to provide an overview on novel fluorescent markers that are used in combination with mitochondrial fusion assays and various live cell microscopy techniques to study mitochondrial dynamics. In particular, approaches to study the movement of mitochondrial proteins and novel imaging techniques (FRET imaging-, 4Pi- and STED-microscopy) that provide high spatial resolution are considered. © 2006 Elsevier B.V. All rights reserved. Keywords: FRAP; FLIP; Fluorescence microscopy; Fusion assay; Mitochondria; Photoactivation; Self labeling protein tag 1. Introduction Next to the mitochondrial dynamics at the level of the whole network, the mitochondrial inner structure is re- Since the identification of mitochondria by electron organized upon various stimuli [4]. Comparatively little data microscopy in the 1950s, it has been become clear that are available on the movement of proteins within mitochondria sometimes mitochondria exist as a single elaborate network and even less is known about the regulation of inner- and at other times they become fragmented [1–3]. -

Effective Axial Resolution in Single-Photon 4Pi Microscopy
4PI CONFOCAL MICROSCOPY Effective Axial Resolution in Single-photon 4Pi Microscopy The feasibility of single-photon 4Pi- confocal microscopy is theoretically demonstrated. By inserting a pair of properly designed multi-ring phase- only pupil filters in the illumination path of a 4Pi microscope the height of the sidelobes of the point spread function is importantly reduced, so that there is not ambiguity in the 3D image. Then, an axial resolution up to four times higher than that of sin- gle-photon confocal microscope can be effectively achieved. Introduction Confocal scanning microscopy is an imaging technique whose main feature is its proverbial optical sectioning capacity [1]. Although confocal technique was specifically designed to improve the axial resolution, it is remarkable that due to the laws of diffraction the resolution in Fig. 1: Schematic lay out of a 4Pi(c)-confocal microscope. Relay lenses are used to focus the pupil filters into the back focal planes of the objectives. the direction of the optical axis is about three times poorer than its lateral coun- terpart. This difference between axial To overcome this problem, it was pro- and lateral resolution leads to ani- posed the use of two-photon excitation sotropic images of three-dimensional [4], which allow an important reduction (3D) specimens. of the height of the secondary axial One attempt to reduce this anisotropy lobes. Then data deconvolution tech- is to insert in the illumination arm of the niques can be used to improve the qual- confocal setup a pupil filter composed by ity of the image [5]. two equal-area transparent rings [2]. -

4Pi Fluorescence Detection and 3D Particle Localization with a Single Objective
4Pi fluorescence detection and 3D particle localization with a single objective J. Schnitzbauer,1 R. McGorty,1 and B. Huang1,* 1Department of Pharmaceutical Chemistry, University of California, 1700 4th St, MC 2532, San Francisco, CA 94158, USA *[email protected] Abstract: Coherent detection through two opposing objectives (4Pi configuration) improves the precision of three-dimensional (3D) single- molecule localization substantially along the axial direction, but suffers from instrument complexity and maintenance difficulty. To address these issues, we have realized 4Pi fluorescence detection by sandwiching the sample between the objective and a mirror, and create interference of direct incidence and mirror-reflected signal at the camera with a spatial light modulator. Multifocal imaging using this single-objective mirror interference scheme offers improvement in the axial localization similar to the traditional 4Pi method. We have also devised several PSF engineering schemes to enable 3D localization with a single emitter image, offering better axial precision than normal single-objective localization methods such as astigmatic imaging. ©2010 Optical Society of America OCIS codes: (180.2520) Fluorescence microscopy; (180.3170) Interference microscopy; (110.6880) Three-dimensional image acquisition; (110.1080) Active or adaptive optics; (350.5730) Resolution. References and links 1. G. J. Schütz, V. Ph. Pastushenko, H. J. Gruber, H.-G. Knaus, B. Pragl, and H. Schindler, “3D Imaging of Individual Ion Channels in Live Cells at 40nm Resolution,” Single Molecules 1(1), 25–31 (2000). 2. M. Speidel, A. Jonás, and E. L. Florin, “Three-dimensional tracking of fluorescent nanoparticles with subnanometer precision by use of off-focus imaging,” Opt. Lett. 28(2), 69–71 (2003). -

Introduction to Superresolution-Zeiss.Pdf
11/15/12 ZEISS Microscopy Online Campus | Introduction to Superresolution Microscopy Contact Us | Carl Zeiss Education in Microscopy and Digital Imaging ZEISS Home ¦ Products ¦ Solutions ¦ Support ¦ Online Shop ¦ ZEISS International ZEISS Campus Home Interactive Tutorials Basic Microscopy Spectral Imaging Spinning Disk Microscopy Optical Sectioning Superresolution Introduction Article Quick Links Introduction Live-Cell Imaging Over the past several decades, fluorescence microscopy has become an Nearfield (NSOM) Fluorescent Proteins essential tool for examining a wide variety of biological molecules, 4Pi and InM pathways, and dynamics in living cells, tissues, and whole animals. In Microscope Light Sources RESOLFT contrast to other techniques (such as electron microscopy), fluorescence Digital Image Galleries imaging is compatible with cells that are being maintained in culture, which PSF Engineering Applications Library enables minimally invasive optical-based observation of events occurring SR-SIM on a large span of timescales. In terms of spatial resolution, several PALM & STORM Reference Library techniques including positron-emission tomography, magnetic resonance Print Version imaging, and optical coherence tomography can generate images of animal and human subjects at resolutions between 10 centimeters and 10 Search micrometers, whereas electron microscopy and scanning probe techniques feature the highest spatial resolution, often approaching the molecular and atomic levels (see Figure 1). Between these two extremes in resolving power lies optical microscopy. Aside from the benefits derived from being able to image living cells, the most significant drawback to all forms of fluorescence microscopy (including widefield, laser scanning, spinning disk, multiphoton, and total internal reflection) are the limits to spatial resolution that were first elucidated and described by Ernst Abbe in the late 1800s.