Aberrations in 4Pi Microscopy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fluorescent Probes for RESOLFT-Type Microscopy at Low Light Intensities
Generation of Novel Photochromic GFPs: Fluorescent Probes for RESOLFT-type Microscopy at Low Light Intensities Dissertation for the award of the degree “Doctor rerum naturalium” Division of Mathematics and Natural Sciences of the Georg-August-Universität Göttingen submitted by Tim Grotjohann from Hannover Göttingen, 2012 Thesis committee members: - Prof. Dr. Stefan Jakobs (1st Referee) Max Planck Institute for Biophysical Chemistry, Göttingen Department of NanoBiophotonics Mitochondrial Structure and Dynamics Group - Prof. Dr. Kai Tittmann (2nd Referee) Georg August University Göttingen Albrecht von Haller Institute Department of Bioanalytics - Dr. Dieter Klopfenstein Georg August University Göttingen Third Institute of Physics Department of Biophysics Date of oral examination: 18th April, 2012 Affidavit I hereby ensure that the presented thesis “Generation of novel photochromic GFPs: fluorescent probes for live cell RESOLFT-type microscopy at low light intensities” has been written independently and with no other sources and aids than quoted. ___________________________________ (Tim Grotjohann) 10th February 2012, Göttingen Table of Contents I. Summary ............................................................................ 4 II. Introduction ........................................................................ 5 1. Fluorescence Microscopy ................................................................................................ 6 a. Fluorescence .............................................................................................................. -

Arrangement of a 4Pi Microscope for Reducing the Confocal Detection
Arrangement of a 4Pi microscope for reducing the confocal detection volume with two-photon excitation Nicolas Sandeau∗and Hugues Giovannini† Received 16 November 2005; received in revised form 7 February 2006; accepted 8 February 2006 Institut Fresnel, UMR 6133 CNRS, Universit´ePaul C´ezanne Aix-Marseille III, F-13397 Marseille cedex 20, France The main advantage of two-photon fluorescence confocal microscopy is the low absorption obtained with live tissues at the wavelengths of operation. However, the resolution of two-photon fluorescence confocal microscopes is lower than in the case of one-photon excitation. The 4Pi microscope type C working in two-photon regime, in which the excitation beams are coherently superimposed and, simultaneously, the emitted beams are also coherently added, has shown to be a good solution for increasing the resolution along the optical axis and for reducing the amplitude of the side lobes of the point spread function. However, the resolution in the transverse plane is poorer than in the case of one-photon excitation due to the larger wavelength in- volved in the two-photon fluorescence process. In this paper we show that a particular arrangement of the 4Pi microscope, referenced as 4Pi’ microscope, is a solution for obtaining a lateral resolution in the two-photon regime sim- ilar or even better to that obtained with 4Pi microscopes working in the arXiv:physics/0610036v1 [physics.bio-ph] 6 Oct 2006 one-photon excitation regime. Keywords: Resolution; Fluorescence microscopy; 4Pi microscopy; Confocal; Detection volume; Two-photon excitation ∗E-mail: [email protected] †E-mail: [email protected]; Tel: +33 491 28 80 66; Fax: +33 491 28 80 67 1 1 Introduction Strong efforts have been made in the last decade to improve the resolution of fluorescence microscopes. -

Photocontrollable Fluorescent Proteins for Superresolution Imaging
BB43CH14-Lippincott-Schwartz ARI 19 May 2014 15:54 Photocontrollable Fluorescent Proteins for Superresolution Imaging Daria M. Shcherbakova,1,2 Prabuddha Sengupta,3 Jennifer Lippincott-Schwartz,3 and Vladislav V. Verkhusha1,2 1Department of Anatomy and Structural Biology, and 2Gruss-Lipper Biophotonics Center, Albert Einstein College of Medicine, Bronx, New York 10461; email: [email protected] 3Section on Organelle Biology, Cell Biology and Metabolism Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland 20892; email: [email protected] Annu. Rev. Biophys. 2014. 43:303–29 Keywords The Annual Review of Biophysics is online at PALM, RESOLFT, PAGFP, PAmCherry, EosFP biophys.annualreviews.org This article’s doi: Abstract 10.1146/annurev-biophys-051013-022836 Superresolution fluorescence microscopy permits the study of biological Copyright c 2014 by Annual Reviews. processes at scales small enough to visualize fine subcellular structures All rights reserved that are unresolvable by traditional diffraction-limited light microscopy. Many superresolution techniques, including those applicable to live cell imaging, utilize genetically encoded photocontrollable fluorescent proteins. Annu. Rev. Biophys. 2014.43:303-329. Downloaded from www.annualreviews.org The fluorescence of these proteins can be controlled by light of specific wavelengths. In this review, we discuss the biochemical and photophysi- by Yeshiva University - Albert Einstein College of Medicine on 06/06/14. For personal use only. cal properties of photocontrollable fluorescent proteins that are relevant to their use in superresolution microscopy. We then describe the recently developed photoactivatable, photoswitchable, and reversibly photoswitch- able fluorescent proteins, and we detail their particular usefulness in single- molecule localization–based and nonlinear ensemble–based superresolution techniques. -

Cooperative 4Pi Excitation and Detection Yields Sevenfold Sharper Optical Sections in Live-Cell Microscopy
4146 Biophysical Journal Volume 87 December 2004 4146–4152 Cooperative 4Pi Excitation and Detection Yields Sevenfold Sharper Optical Sections in Live-Cell Microscopy Hilmar Gugel,* Jo¨rg Bewersdorf,y Stefan Jakobs,y Johann Engelhardt,z Rafael Storz,* and Stefan W. Hellyz *Leica Microsystems Heidelberg GmbH, 68165 Mannheim, Germany; yMax Planck Institute for Biophysical Chemistry, Department of NanoBiophotonics, 37070 Go¨ttingen, Germany; and zGerman Cancer Research Center, High Resolution Optical Microscopy Division, 69120 Heidelberg, Germany ABSTRACT Although the addition of just the excitation light field at the focus, or of just the fluorescence field at the detector is sufficient for a three- to fivefold resolution increase in 4Pi-fluorescence microscopy, substantial improvements of its optical properties are achieved by exploiting both effects simultaneously. They encompass not only an additional expansion of the optical bandwidth, but also an amplified transfer of the newly gained spatial frequencies to the image. Here we report on the realization and the imaging properties of this 4Pi microscopy mode of type C that also is the far-field microscope with the hitherto largest aperture. We show that in conjunction with two-photon excitation, the resulting optical transfer function displays a sevenfold improvement of axial three-dimensional resolution over confocal microscopy in aqueous samples, and more importantly, a marked transfer of all frequencies within its inner region of support. The latter is present also without the confocal pinhole. Thus, linear image deconvolution is possible both for confocalized and nonconfocalized live-cell 4Pi imaging. Realized in a state-of-the-art scanning microscope, this approach enables robust three-dimensional imaging of fixed and live cells at ;80 nm axial resolution. -

Dual-Color 4Pi-Confocal Microscopy with 3D-Resolution in the 100 Nm Range
Ultramicroscopy 90 (2002) 207–213 Dual-color 4Pi-confocal microscopy with 3D-resolution in the 100 nm range Hiroshi Kano, Stefan Jakobs, Matthias Nagorni, Stefan W. Hell* High Resolution Optical Microscopy Group, Max-Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37070 Gottin. gen, Germany Received 21 November 2000; received in revised form 2 July 2001 Abstract We report the development of simultaneous two-color channel recordingin 4Pi-confocal microscopy. A marked increase of spatial resolution over confocal microscopy becomes manifested in 4Pi-confocal three-dimensional (3D) data stacks of dual-labeled objects. The fundamentally improved resolution is verified both with densely labeled fluorescence beads as well as with membrane labeled fixed Escherichia coli. The synergistic combination of dual-color 4Pi-confocal recording with image restoration results in dual-color imaging with a 3D resolution in the 100 nm range. r 2002 Elsevier Science B.V. All rights reserved. 1. Introduction the typically 3.5 times sharper main maximum [1]. The side-lobes are removed mathematically by a By providinga fundamental improvement of linear filter, in which case a typical axial resolution axial sectioning, 4Pi-confocal microscopy [1] is a of 130–140 nm is achieved at a two-photon promisingdevelopment in three-dimensional (3D) excitation wavelength of 700–800 nm [3]. Alterna- far-field fluorescence microscopy. The 4Pi-confo- tively, the effective 3D-point spread function cal microscope owes its superior sectioningcap- (PSF) of the 4Pi-confocal microscope is used ability to the coherent addition of the spherical altogether to restore the 4Pi-confocal image data wavefronts produced by two opposingobjective with a non-linear restoration algorithm [4]. -

Two-Photon Excitation and Photoconversion of Eosfp in Dual-Color 4Pi Confocal Microscopy
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biophysical Journal Volume 92 June 2007 4451–4457 4451 Two-Photon Excitation and Photoconversion of EosFP in Dual-Color 4Pi Confocal Microscopy Sergey Ivanchenko,* Sylvia Glaschick,* Carlheinz Ro¨cker,* Franz Oswald,y Jo¨rg Wiedenmann,z and G. Ulrich Nienhaus*§ *Institute of Biophysics, yDepartment of Internal Medicine I, and zInstitute of General Zoology and Endocrinology, University of Ulm, 89069 Ulm, Germany; and §Department of Physics, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 ABSTRACT Recent years have witnessed enormous advances in fluorescence microscopy instrumentation and fluorescent marker development. 4Pi confocal microscopy with two-photon excitation features excellent optical sectioning in the axial di- rection, with a resolution in the 100 nm range. Here we apply this technique to cellular imaging with EosFP, a photoactivatable autofluorescent protein whose fluorescence emission wavelength can be switched from green (516 nm) to red (581 nm) by irradiation with 400-nm light. We have measured the two-photon excitation spectra and cross sections of the green and the red species as well as the spectral dependence of two-photon conversion. The data reveal that two-photon excitation and photo- activation of the green form of EosFP can be selectively performed by choosing the proper wavelengths. Optical highlighting of small subcellular compartments was shown on HeLa cells expressing EosFP fused to a mitochondrial targeting signal. After three-dimensionally confined two-photon conversion of EosFP within the mitochondrial networks of the cells, the converted re- gions could be resolved in a 3D reconstruction from a dual-color 4Pi image stack. -

Two Approaches for a Simpler STED Microscope Using a Dual-Color Laser Or a Single Wavelength Ancuţa Teodora Şcheul
Two approaches for a simpler STED microscope using a dual-color laser or a single wavelength Ancuţa Teodora Şcheul To cite this version: Ancuţa Teodora Şcheul. Two approaches for a simpler STED microscope using a dual-color laser or a single wavelength. Other [cond-mat.other]. Université de Grenoble, 2013. English. NNT : 2013GRENY040. tel-01558306 HAL Id: tel-01558306 https://tel.archives-ouvertes.fr/tel-01558306 Submitted on 7 Jul 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THESE` Pour obtenir le grade de DOCTEUR DE L’UNIVERSITE´ DE GRENOBLE Specialit´ e´ : Optique, Nanophysique Arretˆ e´ ministeriel´ : 7 aoutˆ 2006 Present´ ee´ par S¸cheul Ancut¸aTeodora These` dirigee´ par Jean-Claude Vial et codirigee´ par Irene` Wang prepar´ ee´ au sein du Laboratoire interdisciplinaire de Physique et de l’Ecole Doctorale de Physique, Grenoble Two approaches for a simpler STED microscope using a dual- color laser or a single wavelength These` soutenue publiquement le 22 Novembre 2013, devant le jury compose´ de : Dominique Bourgeois Directeur de Recherche -

Super-Resolution Microscopy 1
Semrock White Paper Series: Super-resolution Microscopy 1. Introduction Fluorescence microscopy has revolutionized the study of biological samples. Ever since the invention of fluorescence microscopy towards the beginning of the 20th century, significant technological advances have enabled elucidation of biological phenomenon at cellular, sub- cellular and even at molecular levels. However, the latest incarnation of the modern fluorescence microscope has led to a paradigm shift. This wave is about breaking the diffraction limit first proposed in 1873 by Ernst Abbe. The implications of this development are profound. This new technology, called super-resolution microscopy, allows for the visualization of cellular samples with a resolution similar to that of an electron microscope, yet it retains the advantages of an optical fluorescence microscope. This means it is possible to uniquely visualize desired molecular species in a cellular environment, even in three dimensions and now in live cells – all at a scale comparable to the spatial dimensions of the molecules under investigation. This article provides an overview of some of the key recent developments in super-resolution microscopy. 2. The problem So what is wrong with a conventional fluorescence microscope? To understand the answer to this question, consider a Green Fluorescence Protein (GFP) molecule that is about 3 nm in diameter and about 4 nm long. When a single GFP molecule is imaged using a 100X objective the size of the image should be 0.3 to 0.4 microns (100 times the size of the object). Yet the smallest spot that can be seen at the camera is about 25 microns. Which corresponds to an object size of about 250 nm. -
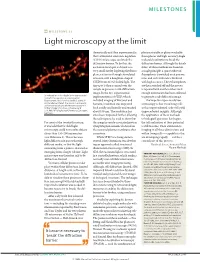
Milestone 21: Light Microscopy at the Limit
MILESTONES MILESTONE 21 Light microscopy at the limit theoretically and then experimentally, photoactivatable or photoswitchable that a stimulated-emission-depletion fluorophores and high-accuracy single- (STED) microscope can break the molecule localization to break the diffraction barrier. To do this, the diffraction barrier. Although the details excitation focal spot is shrunk to a differ, all three methods are based on very small size by depleting the fluoro- a single principle: a sparse subset of phores at its rim through stimulated fluorophores is switched on at any one emission with a doughnut-shaped time, and each molecule is localized STED beam of red-shifted light. The with high accuracy. These fluorophores tiny spot is then scanned over the are then switched off and the process sample to generate a sub-diffraction is repeated with another subset until image. In the first experimental enough information has been collected Stimulated-emission-depletion image recorded from a living neuron of an organotypical implementation of STED, which to generate a sub-diffraction image. hippocampal slice, showing dendritic spines in included imaging of live yeast and The hope for super-resolution unprecedented detail. The neuron is expressing yellow fluorescent protein. Image courtesy of K. bacteria, resolution was improved microscopy is that visualizing cells Willig, V. Nägerl, N. Urban, T. Bonhoeffer & both axially and laterally and reached at this unprecedented scale will yield S. W. Hell, MPI Biophysical Chemistry, Göttingen, Germany about 100 nm. The resolution has unprecedented insights. Although since been improved further, allowing the application of these methods the technique to be used to show that to biological questions has begun, For most of the twentieth century, the synaptic vesicle-associated protein the full realization of their potential it was held that far-field light synaptotagmin remains clustered on is still to come. -

Accurate 4Pi Single-Molecule Localization Using an Experimental PSF Model
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.18.997163; this version posted March 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Accurate 4Pi single-molecule localization using an experimental PSF model Yiming Li1,2, Elena Buglakova2,3, Yongdeng Zhang4, Jervis Vermal Thevathasan2,5, Joerg Bewersdorf4,6,7,8, Jonas Ries2,* 1Department of Biomedical Engineering, Southern University of Science and Technology, Shenzhen 518055, China 2European Molecular Biology Laboratory, Cell Biology and Biophysics, Heidelberg 69117, Germany 3Moscow Institute of Physics and Technology, Dolgoprudny 141701, Russia 4Department of Cell Biology, Yale School of Medicine, New Haven, CT 06511, USA 5Faculty of Biosciences, Heidelberg University, Heidelberg 69120, Germany 6Kavli Institute for Neuroscience, Yale School of Medicine, New Haven, CT 06511, USA 7Nanobiology Institute, Yale University, West Haven, CT 06516, USA 8Department of Biomedical Engineering, Yale University, New Haven, CT 06511, USA *Correspondence should be addressed to J.R. ([email protected]) Interferometric single-molecule localization microscopy (iPALM, 4Pi-SMS) uses multiphase interferometry to localize single fluorophores and achieves nanometer isotropic resolution in 3D. The current data analysis workflow, however, fails to reach the theoretical resolution limit due to the suboptimal localization algorithm. Here, we develop a method to fit an experimentally derived point spread function (PSF) model to the interference 4Pi-PSF. As the interference phase is not fixed with respect to the shape of the PSF, we decoupled the phase term in the model from the 3D position of the PSF. -

RESOLFT Light-Sheet Microscopy
Dissertation submitted to the Combined Faculties for the Natural Sciences and for Mathematics of the Ruperto-Carola University of Heidelberg, Germany for the degree of Doctor of Natural Sciences Put forward by Diplom-Physiker Patrick Hoyer Born in Weiden i. d. OPf. Oral examination: December 17, 2014 RESOLFT Light-sheet Microscopy Referees: Prof. Dr. Stefan W. Hell Prof. Dr. Wolfgang Petrich «φύσις κρύπτεσθαι φιλεῖ.» (“Nature loves to hide.”) Heraklit, Fragmentum, B 123. Abstract e concept of RESOLFT was introduced to break the resolution limit in fluo- rescence microscopy, which is set by the physical phenomenon of diffraction. It enables the separation of fluorophores inside a focal volume by driving re- versible optical transitions between two discernible states. e family of re- versibly switchable fluorescent proteins (RSFPs) with long-lived on- and off- states allows for RESOLFT imaging at low light levels. Up to now, RESOLFT involving RSFPs as fluorescent markers has been exclusively demonstrated to enhance the resolution in the lateral dimension. In this work, a novel RSFP- based RESOLFT microscope is presented, that, for the first time, breaks the diffraction barrier in the axial direction by switching fluorophores in the vol- ume of a light-sheet. It is realized with the optical arrangement of a selec- tive plane illumination microscope (SPIM), that illuminates only a thin sec- tion of a sample perpendicular to the detection axis and thus reduces the overall light exposure in volume recordings. e symbiotic combination of RSFP-based RESOLFT and SPIM, the so called RESOLFT-SPIM nanoscope, offers highly parallelized, fast imaging of living biological specimens with low light doses and sub-diffraction axial resolution. -

Experimental Stochastics in High-Resolution Fluorescence
High-Resolution Fluorescence Microscopy with Photoswitchable Fluorescent Proteins Dissertation zur Erlangung des mathematisch-naturwissenschaftlichen Doktorgrades ”Doctor rerum naturalium” der Georg-August-Universitat¨ Gottingen¨ vorgelegt von Hannes Bock aus Hamburg Gottingen,¨ 2008 . D 7 Referent: Prof. Dr. M. Munzenberg¨ Koreferent: Prof. Dr. S. W. Hell Tag der mundlichen¨ Prufung:¨ High-Resolution Fluorescence Microscopy with Photoswitchable Fluorescent Proteins Summary The diffraction limit in far-field fluorescence microscopy can be overcome by photoswitch- ing the marker molecules between a fluorescent and a non-fluorescent state. Photoswitching enables the successive generation and readout of nano-sized fluorescent areas in the sample. Subsequently, these areas can be assembled to form a super-resolution image of the speci- men. For the first time in this work, one and the same molecular switching mechanism of a reversibly switchable fluorescent protein (RSFP) was used to realize two alternative varia- tions of this general nanoscopy principle: one based on the targeted switching of ensembles of molecules and the other based on the stochastic switching of single emitters. RSFPs are of particular interest for these techniques because they allow the non-invasive study of pro- cesses in living cells and can be efficiently switched using low light intensities, thus minimiz- ing photo stress and possible photo-induced damage of the cell. Furthermore, the protein’s photoswitching properties can be improved via targeted mutagenesis. In the case of both nanoscopy techniques presented here, image acquisition could be significantly improved and accelerated as compared to related approaches. For the first time, two-color far-field fluores- cence nanoscopy based on the photoswitching of single emitters was successfully realized in biological samples.