Tfri-Ontario Node Research Symposium Thursday December 12, 2019
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Canada Scientific Advisory Committee (CSAC)
Canada Scientific Advisory Committee (CSAC) Alan Bernstein, PhD – Co-chairperson Canadian Institute for Advanced Research Toronto, Ontario Alan Bernstein is President of the Canadian Institute for Advanced Research (CIFAR), Canada’s global research institute. From 2008- 2011, Bernstein was the executive director of the Global HIV Vaccine Enterprise, an international alliance of researchers and funders charged with accelerating the search for an HIV vaccine. Previously, he served as the founding president of the Canadian Institutes of Health Research (2000-2007), Canada’s federal agency for the support of health research. In that capacity, he led the transformation of health research in Canada. After receiving his PhD from the University of Toronto, and following postdoctoral work in London, Bernstein joined the Ontario Cancer Institute (1974-1985). In 1985, he joined the new Samuel Lunenfeld Research Institute in Toronto, was named Associate Director in 1988 and then Director of Research (1994-2000). Internationally known for his contributions to our understanding of the molecular basis of cancer, Bernstein has made extensive contributions to the study of stem cells, hematopoiesis and cancer. He chairs or is a member of advisory and review boards in Canada, the US, UK and Italy. Bernstein has received numerous awards and honourary degrees for his contributions to science, including the 2008 Gairdner Wightman Award, induction into the Canadian Medical Hall of Fame, and the Henry G. Friesen International Prize in Health Research. He is a Senior Research Fellow of Massey College, received the Order of Ontario in 2018 and was appointed an Officer of the Order of Canada in 2002. -

ANNUAL REPORT 2015/2016 Message from the Minister of Research, Innovation and Science
ANNUAL REPORT 2015/2016 Message from the Minister of Research, Innovation and Science On behalf of the Government of Ontario I am pleased to have the opportunity to extend my thanks to the Ontario Institute for Cancer Research (OICR) for another successful year in its progress to meet the cancer challenge. A report issued recently by Cancer Care Ontario tells us that one in two Ontarians will develop cancer in their lifetime and cancer is the leading cause of death in the province. We have to do everything possible to alleviate the burden of cancer on Ontario families by improving the prevention, detection, diagnosis and treatment of cancer. We are proud of our investment in OICR over the last 10 years and recently approved the Institute’s Strategic Plan for 2016-2021. OICR has demonstrated leadership in the cancer community not only in its research programs but also in its focus on moving discoveries into the clinic. An example is the development of a novel oncolytic viral immunotherapy which is now in clinical trials. Through OICR’s commercialization partner, the Fight Against Cancer Innovation Trust, financial and in-kind support are attracted which makes it possible to accelerate development and therefore bring new hope to patients. The past year has been one of accomplishment and one of transition. Dr. Calvin Stiller, who was instrumental in the creation of the Institute, has stepped down as Chair of the Board of Directors. We are most grateful for his vision and leadership. Dr. Tom Hudson, who built the Institute, attracted talented researchers and launched provincial, national and international initiatives that are helping to make Ontario a world leader in cancer research, has moved on to the next phase of his career. -

Princess Margaret Cancer Centre Annual Report
2 015 A NNU A L REPORT 3 Leadership Message 4 Our Program 4 Who we are Welcome to the 2015 Annual Report for the Princess Margaret Cancer Centre at the University Health Network (UHN). This year proved to be both exciting and engaging as we continue to deliver revolutionary cancer care for our patients, and we are looking 6 Clinical Care forward to sharing the progress we have made. The 2015 edition showcases the recent 6 Our Clinical Programs LEADERSHIP activities and accomplishments of our people, departments, disease groups, and research and education programs. As one of the largest comprehensive cancer treatment facilities in the world, we have a great deal to share as we continue our efforts to be on the 8 Personalized Cancer Medicine MESSAGE frontiers of medical, surgical and radiation oncology, embracing the latest technology 8 Our Strategy and international best-practices, and setting standards for patient care. 9 We Are Transforming Patient Care In 2015, we celebrated 20 years of our presence on University Avenue; the hub of the discovery district. The move not only signified our commitment to meet the increased 11 We Are Augmenting Correlative Cancer Biology demands and evolving needs of our patients, but also encouraged collaboration, innovation, and research, enabling us to continue making progress in conquering cancer. 12 We Are Accelerating Guided Therapeutics Today, we again embrace change as we drive implementation of our space plan, with Marnie Escaf MHA, HBBA 15 We Are Expanding Novel Therapeutics Senior Vice President a focus on redeveloping facilities to improve the patient experience, including patient Executive Lead amenities, access, flow, and wayfinding. -

(DMOH) Toronto General Hospital, University Health Network
General & Consultative Academic Hematologist, Division of Medical Oncology and Hematology (DMOH) Toronto General Hospital, University Health Network The Department of Medicine, Faculty of Medicine at the University of Toronto and the Division of Medical Oncology and Hematology at the University Health Network (UHN) are seeking to recruit a general academic hematologist for the Toronto General Hospital’s Blood Disorders Program. The successful candidate will have an academic position of Clinician Investigator and must be eligible for a full-time clinical academic appointment at the rank of Lecturer or Assistant Professor at the University of Toronto. Effective start date is September 1, 2017, or shortly thereafter. The position is well suited to early career physicians with an interest in non- and pre-malignant hematology. Strong collaborative skills are required as there is an opportunity to form closer links with the malignant hematology clinics and with our principal referring internal stakeholders. The role will be primarily ambulatory clinic based, serving hematological complications from UHN’s Cardiac, Multi-Organ Transplant and Internal Medicine programs, as well as the local community. The candidate should have a strong academic interest in postgraduate MD teaching or in quality improvement/patient safety and uphold standards of excellence. Additional qualifications in these domains are preferred. The Blood Disorders Program is a core program within the Division of Medical Oncology & Hematology at the University Health Network. As one of the top health networks in the world, the University Health Network encompasses both the Princess Margaret Cancer Centre as well as the Toronto General Hospital along with each of their respective world-class research institutes: The Ontario Cancer Institute and Toronto General Research Institute. -

Scientific Advisory Committee on Oncology Therapies (SAC-OT)
Scientific Advisory Committee on Oncology Therapies (SAC-OT) Membership List Core Members Alexander H.G. Paterson, MD, FRCP, FACP, MBChB (Chair) Medical Oncologist, Department of Medical Oncology, Tom Baker Cancer Centre Professor, Department of Medicine and Oncology, University of Calgary Calgary, Alberta Biography: Alexander Paterson graduated Medicine from Edinburgh University, United Kingdom in 1977 and subsequently trained at St. Bartholomew’s Hospital and Royal Marsden, London, England. He is a Fellow of the Royal College of Physicians and has been a Medical Oncologist at the Tom Baker Cancer Centre in Calgary since 1990. He is also Professor in the Departments of Medicine and Oncology at the University of Calgary, Alberta since 1995. He is a member of the Alberta Out-of-Province/Country Health Services Appeal Panel and the Board of Directors of the National Surgical Adjuvant Breast and Bowel Project (NSAPB). He is also Chair of the NSABP Protocol B-34 and the Alberta Breast Cancer Programme. He has published over 100 articles, authored over 15 book chapters, is the Editor of Fundamental Problems in Breast Cancer (I and II) and contributes a regular column to Alberta Doctors’ Digest. He has given over 200 invited lectures. Rick Abbott, BScPharm, RPEBC Pharmacy Manager, Provincial Systemic Therapy, Dr. H. Bliss Murphy Cancer Centre, Eastern Health St. John’s, Newfoundland and Labrador Biography: Rick Abbott graduated from the School of Pharmacy in 1990 at the Memorial University of Newfoundland, St. John’s, Newfoundland and Labrador. He has been the Pharmacy Manager of the Provincial Systemic Therapy since 2002 and is actively involved in the Pharmacy Profession. -

Annual Report 2010-2011
ON TARIO IN STIT U T E FOR C A N C E R RE S Ontario Institute for E ARC H ANNU Cancer Research A L REP ANNUAL REPORT 2010/11 ORT 2010/11 MaRS Centre, South Tower 101 College Street Suite 800 Toronto, Ontario Canada M5G 0A3 Telephone 416.977.7599 Toll-free 1.866.678.6427 [email protected] www.oicr.on.ca Funding for the Ontario Institute for Cancer Research is provided by the Government of Ontario. CJ23389 Cover.indd 1 11-07-07 6:56 PM Message from the Minister of Research and Innovation It is my pleasure to thank the Ontario Institute for Cancer Research (OICR) for the outstanding work it has done over For information about the the past year to improve the health and wellbeing of the people of Ontario. Ontario Institute for Cancer Research please contact: T he work you do has a direct impact on the lives of everyone in the province. It offers hope to the nearly 70,000 Ontarians Rhea Cohen who will be diagnosed with cancer in 2011 and to many millions more around the world. T he survival rate for most types Director of Communications [email protected] of cancer is constantly improving – and your work is helping to make that happen. 416-673-6642 OICR is recognized around the world for its leadership in cancer research. T he Institute’s high profile role – and that of its President and Scientific Director, Dr. Tom Hudson – in the International Cancer Genome Consortium is just one example of that leadership. -
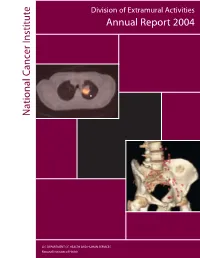
National C Ancer Institute Annual Report 2004
Division of Extramural Activities Annual Report 2004 National Cancer Institute National Cancer U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES National Institutes of Health Molecular Imaging for Cancer The NCI has made a significant committment to support the discovery, development, and delivery of cutting- edge molecular imaging agents and technologies. The field of molecular imaging has grown rapidly in response to this committment, and the applications being developed will have a far-reaching impact on the detection, diagnosis, and treatment of cancer patients. The images on the cover demonstrate the power of two different types of molecular imaging techniques applied to cancer detection. The image on the top is generated using positron emission tomography-computed tomog- raphy (PET-CT) fusion imaging (image reproduced with permission from David W. Townsend, Ph.D., Department of Radiology, University of Tennessee). Computed-tomography (CT) alone, a technique based on the use of x-rays, is capable of generating images with exquisite anatomical detail, but does not provide the clinician with physiologic information about lesions that are detected. Conversely, positron emission tomogra- phy (PET) alone, a technique based on the use of radiolabeled imaging agents, provides the clinician with func- tional information about whether a lesion is cancerous or not, but the resulting images do not contain the anatomic detail necessary to determine exact location within the body. In the image shown, the radiolabeled agent being used is 2-[18F]fluoro-2-deoxy-D-glucose ([18F] FDG), an agent that reports on the increased metabolism occuring in a cancer cell. In the past, these two types of images were acquired using two sepa- rate machines at two different times. -

STEMCELLNETWORK.CA the Mission of the Stem Cell
STEM CELL NETWORK SUMMER 2006 VOLUME 5, NUMBER 1 FATHERS OF THE FIELD How two quiet Canadians changed the course of history in biological research. CRITICAL MASS AHEAD OF THE CURVE A CANADIAN A pioneering past and a culture of Finding the way inside the ‘black box’ COMES HOME collaboration combine to make Toronto of cancer, Dr. John Dick has changed One of America’s leading researchers a world leader in stem cell science. our understanding of how to fight takes up a new challenge in the city the deadly disease. where he began his brilliant career. WWW.STEMCELLNETWORK.CA The mission of the Stem Cell Network is to be a catalyst for realizing the full potential of stem cell research for Canadians. STEM CELL NETWORK Frank Gleeson, Chair, Board of Directors Dr. Michael Rudnicki, Scientific Director Dr. Janet Rossant, Deputy Scientific Director Drew Lyall, Executive Director Cathy Campbell, SCN Communications Lori Barron, SCN Communications Joe Sornberger, Writer CONTACT US AT: 451 Smyth Road, Ottawa, ON K1H 8M5 Tel: (613)562.5696 Fax: (613)562.5631 Website: www.stemcellnetwork.ca Publication Mail Agreement Number 40664504 The contents of this publication may be reprinted or used in radio or television without permission. However, a credit is requested. In print, please send a copy to the Stem Cell Network. STEM CELL NETWORK TABLE OF CONTENTS Welcome to Toronto - Dr. Michael Rudnicki .......................................1 Critical Mass .................................................................................2 The Fathers of the Field .................................................................8 A Canadian comes home - Interview with Dr. Gordon Keller ...............14 The world’s best feel right at home ..............................................15 On the Cover: Ahead of the curve - Interview with Dr. -

OICR Strategic Plan 2010-2015 Was Generated in Parallel with the Pan-Canadian Cancer Research Strategy 2010-2014 and the National Breast Cancer Research Framework
Strategic Plan 2010-2015 OICR timeline from 2000 to 2010 The Government of The Government of Ontario Ontario asked Dr. Cabinet approved created the Ontario Cancer OCRN’s board of Calvin Stiller to lead a the Stiller Committee Research Network (OCRN), a directors held its group to recommend recommendation to three-year initiative with three OCRN was first meeting. how a $50 million focus on translational components: The Government of incorporated Mr. George Glover investment in cancer research related to Cancer Research Fund Ontario hired as a federal was appointed the research could have development of new Virtual Information Network Dr. Robert Phillips to not-for-profit first chair of the the greatest impact. cancer therapies. Tumour Bank Network. set up and run OCRN. corporation. board of directors. 05 . 2000 12 . 2000 11 . 2001 09 . 2000 09 . 2001 01 . 2002 Dr. Bette Stephenson The Ontario Innovation Trust provided The Government of Ontario Drs. funding to explore what it would announced it would spend Stephenson The Government of take for Ontario to make a significant $1 billion over 10 years to and Stiller The Cancer Ontario announced impact on cancer; Mr. George Connell, create a cancer research Dr. John Evans submitted Research $50 million in Dr. Louis Siminovitch, Dr. Charles institute; Drs. Bette was appointed their report Fund held additional funding for Hollenberg, Ms. Michele Noble and Stephenson and Calvin Stiller chair of OCRN’s to the its first grant OCRN, extending its Dr. Patrick Lafferty began work on a were asked to spearhead the board of Government competition. mandate to four years. -

Retraction Watch
Retraction Watch Tracking retractions as a window into the scientific process Author asks to retract nearly 20-year old paper over figure questions, lack of data with 22 comments The last author of a 1999 paper has asked the journal to retract it less than one month after a user raised questions about images on PubPeer. Yesterday, last author Jim Woodgett posted a note on the site saying the author who generated the figures in question could not find the original data, and since he agreed the images appeared “suspicious,” he had contacted the journal to retract the paper. Here’s the note from Woodgett, based at Lunenfeld-Tanenbaum Research Institute at Mount Sinai Hospital in Toronto: …the person who generated the original data cannot source it and, as a consequence, a request to retract this paper based on the discrepancies in figure 5B and C has been submitted and approved. The PubPeer exchange is over a pair of figures in the 1999 paper, “Regulation of the protein kinase activity of Shaggy(Zeste-white3) by components of the wingless pathway in Drosophila cells and embryos,” which has been cited 77 times, according to Thomson Reuters Web of Science. One day after a user posted the figures, Woodgett responded: I’m senior author on that paper and those figures were all from my lab and most likely generated by the first author, Dr. Laurent Ruel, who is a fantastic geneticist. I’ll ask him to dig out the primary auto rads (hopefully still has them from 17 years ago) but looks at my first glance like noise (the placement is just bizarre for any malicious intent, no?). -

James E. Till Fonds B2005-0031 B2010-0028
University of Toronto Archives and Records Management Services James E. Till fonds B2005-0031 B2010-0028 Harold Averill, July 2006 Revised by Garron Wells, January 2011 Emily Sommers, April 2021 © University of Toronto Archives and Records Management Services, 2021 James E. Till fonds University of Toronto Archives B2005-0031, B2010-0028 Contents Biographical note ..................................................................................................................... 3 Scope and content ................................................................................................................... 5 Series 1: Personal ...................................................................................................................... 6 Series 2: Correspondence ....................................................................................................... 7 Series 3: University of Toronto .................................................................................................. 8 Series 4: National Cancer Institute of Canada, Ontario Cancer Institute ....................... 10 Series 5: Professional organizations ...................................................................................... 12 Series 6: Research ................................................................................................................... 13 Series 7: Manuscripts and publications ............................................................................... 14 Series 8: Addresses ................................................................................................................ -

Inhibition of Mitochondrial Translation As a Therapeutic Strategy
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Cancer Cell Article Inhibition of Mitochondrial Translation as a Therapeutic Strategy for Human Acute Myeloid Leukemia Marko Skrtic, 1 Shrivani Sriskanthadevan,1 Bozhena Jhas,1 Marinella Gebbia,2 Xiaoming Wang,1 Zezhou Wang,1 Rose Hurren,1 Yulia Jitkova,1 Marcela Gronda,1 Neil Maclean,1 Courteney K. Lai,3 Yanina Eberhard,1 Justyna Bartoszko,1 Paul Spagnuolo,1 Angela C. Rutledge,1 Alessandro Datti,4,5 Troy Ketela,2 Jason Moffat,2 Brian H. Robinson,6 Jessie H. Cameron,6 Jeffery Wrana,4 Connie J. Eaves,3 Mark D. Minden,1 Jean C.Y. Wang,1,7 John E. Dick,7 Keith Humphries,3 Corey Nislow,2 Guri Giaever,2 and Aaron D. Schimmer1,* 1Campbell Family Cancer Research Institute, Princess Margaret Hospital, Ontario Cancer Institute, Toronto, Ontario M5G 2M9, Canada 2Department of Molecular Genetics, Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, Ontario M5S 3E1, Canada 3Terry Fox Laboratory, British Columbia Cancer Agency, Vancouver, British Columbia V5Z 1L3, Canada 4Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario M5G 1X5, Canada 5Department of Experimental Medicine and Biochemical Sciences, University of Perugia, 06126 Perugia, Italy 6Genetics and Genome Biology, The Research Institute, The Hospital for Sick Children, Toronto, Ontario M5G 1X8, Canada 7Division of Stem Cell and Developmental Biology, Campbell Family Institute for Cancer Research, Ontario Cancer Institute, Toronto, Ontario M5G 1L7, Canada *Correspondence: [email protected] DOI 10.1016/j.ccr.2011.10.015 SUMMARY To identify FDA-approved agents targeting leukemic cells, we performed a chemical screen on two human leukemic cell lines and identified the antimicrobial tigecycline.