Effector T Cells and T Lymphocyte Attenuator-Positive Entry Mediator
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

ENSG Gene Encodes Effector TCR Pathway Costimulation Inhibitory/Exhaustion Synapse/Adhesion Chemokines/Receptors
ENSG Gene Encodes Effector TCR pathway Costimulation Inhibitory/exhaustion Synapse/adhesion Chemokines/receptors ENSG00000111537 IFNG IFNg x ENSG00000109471 IL2 IL-2 x ENSG00000232810 TNF TNFa x ENSG00000271503 CCL5 CCL5 x x ENSG00000139187 KLRG1 Klrg1 x ENSG00000117560 FASLG Fas ligand x ENSG00000121858 TNFSF10 TRAIL x ENSG00000134545 KLRC1 Klrc1 / NKG2A x ENSG00000213809 KLRK1 Klrk1 / NKG2D x ENSG00000188389 PDCD1 PD-1 x x ENSG00000117281 CD160 CD160 x x ENSG00000134460 IL2RA IL-2 receptor x subunit alpha ENSG00000110324 IL10RA IL-10 receptor x subunit alpha ENSG00000115604 IL18R1 IL-18 receptor 1 x ENSG00000115607 IL18RAP IL-18 receptor x accessory protein ENSG00000081985 IL12RB2 IL-12 receptor x beta 2 ENSG00000186810 CXCR3 CXCR3 x x ENSG00000005844 ITGAL CD11a x ENSG00000160255 ITGB2 CD18; Integrin x x beta-2 ENSG00000156886 ITGAD CD11d x ENSG00000140678 ITGAX; CD11c x x Integrin alpha-X ENSG00000115232 ITGA4 CD49d; Integrin x x alpha-4 ENSG00000169896 ITGAM CD11b; Integrin x x alpha-M ENSG00000138378 STAT4 Stat4 x ENSG00000115415 STAT1 Stat1 x ENSG00000170581 STAT2 Stat2 x ENSG00000126561 STAT5a Stat5a x ENSG00000162434 JAK1 Jak1 x ENSG00000100453 GZMB Granzyme B x ENSG00000145649 GZMA Granzyme A x ENSG00000180644 PRF1 Perforin 1 x ENSG00000115523 GNLY Granulysin x ENSG00000100450 GZMH Granzyme H x ENSG00000113088 GZMK Granzyme K x ENSG00000057657 PRDM1 Blimp-1 x ENSG00000073861 TBX21 T-bet x ENSG00000115738 ID2 ID2 x ENSG00000176083 ZNF683 Hobit x ENSG00000137265 IRF4 Interferon x regulatory factor 4 ENSG00000140968 IRF8 Interferon -

The TNF and TNF Receptor Review Superfamilies: Integrating Mammalian Biology
Cell, Vol. 104, 487±501, February 23, 2001, Copyright 2001 by Cell Press The TNF and TNF Receptor Review Superfamilies: Integrating Mammalian Biology Richard M. Locksley,*²³k Nigel Killeen,²k The receptors and ligands in this superfamily have and Michael J. Lenardo§k unique structural attributes that couple them directly to *Department of Medicine signaling pathways for cell proliferation, survival, and ² Department of Microbiology and Immunology differentiation. Thus, they have assumed prominent ³ Howard Hughes Medical Institute roles in the generation of tissues and transient microen- University of California, San Francisco vironments. Most TNF/TNFR SFPs are expressed in the San Francisco, California 94143 immune system, where their rapid and potent signaling § Laboratory of Immunology capabilities are crucial in coordinating the proliferation National Institute of Allergy and Infectious Diseases and protective functions of pathogen-reactive cells. National Institutes of Health Here, we review the organization of the TNF/TNFR SF Bethesda, Maryland 20892 and how these proteins have been adapted for pro- cesses as seemingly disparate as host defense and or- ganogenesis. In interpreting this large and highly active Introduction area of research, we have focused on common themes that unite the actions of these genes in different tissues. Three decades ago, lymphotoxin (LT) and tumor necro- We also discuss the evolutionary success of this super- sis factor (TNF) were identified as products of lympho- familyÐsuccess that we infer from its expansion across cytes and macrophages that caused the lysis of certain the mammalian genome and from its many indispens- types of cells, especially tumor cells (Granger et al., able roles in mammalian biology. -

SLPI and Soluble BTLA As Immunological Markers in Severe Bacterial Infections
SLPI and soluble BTLA as immunological markers in severe bacterial infections To my family Örebro Studies in Medicine 211 ANNA LANGE SLPI and soluble BTLA as immunological markers in severe bacterial infections © Anna Lange, 2020 Title: SLPI and soluble BTLA as immunological markers in severe bacterial infections Publisher: Örebro University 2020 www.oru.se/publikationer Print: Örebro University, Repro 04/2020 ISSN 1652-4063 ISBN 978-91-7529-335-6 Abstract Anna Lange (2020): SLPI and soluble BTLA as immunological markers in severe bacterial infections. Örebro Studies in Medicine 211. Clinical presentation, and outcome of infections are affected by host-, and etiology- (focus of infection and pathogen) related factors. The im- mune response is controlled by a network of regulating pathways. This thesis focuses on Secretory Leukocyte Protease Inhibitor (SLPI), a protease inhibitor with anti-inflammatory properties, and the previously non-studied soluble isoform of B and T lymphocyte attenuator (sBTLA), a membrane-associated regulatory protein. Plasma concentrations of SLPI and sBTLA were assessed in relation to etiology, severity, mortality, and markers of inflammation and immunosuppression, in i) community- acquired pneumonia (CAP) (SLPI), ii) intensive care unit (ICU) treated severe sepsis and septic shock (sBTLA), and iii) dynamically in BSI (SLPI and sBTLA). Main findings were: higher expression of SLPI in pneumonia, com- pared to other sources, higher initial concentrations in Streptococcus pneumoniae, and Staphylococcus aureus BSI, compared to Escherichia coli BSI, and higher SLPI concentrations in sepsis compared to non-septic BSI. Interestingly, men with pneumonia had higher plasma levels of SLPI, both in CAP and BSI. Likewise, sBTLA was associated with severity, but preferentially at higher organ failure scores. -

Human TNFRSF18 ELISA Kit (ARG81453)
Product datasheet [email protected] ARG81453 Package: 96 wells Human TNFRSF18 ELISA Kit Store at: 4°C Component Cat. No. Component Name Package Temp ARG81453-001 Antibody-coated 8 X 12 strips 4°C. Unused strips microplate should be sealed tightly in the air-tight pouch. ARG81453-002 Standard 2 X 10 ng/vial 4°C ARG81453-003 Standard/Sample 30 ml (Ready to use) 4°C diluent ARG81453-004 Antibody conjugate 1 vial (100 µl) 4°C concentrate (100X) ARG81453-005 Antibody diluent 12 ml (Ready to use) 4°C buffer ARG81453-006 HRP-Streptavidin 1 vial (100 µl) 4°C concentrate (100X) ARG81453-007 HRP-Streptavidin 12 ml (Ready to use) 4°C diluent buffer ARG81453-008 25X Wash buffer 20 ml 4°C ARG81453-009 TMB substrate 10 ml (Ready to use) 4°C (Protect from light) ARG81453-010 STOP solution 10 ml (Ready to use) 4°C ARG81453-011 Plate sealer 4 strips Room temperature Summary Product Description ARG81453 Human TNFRSF18 ELISA Kit is an Enzyme Immunoassay kit for the quantification of Human TNFRSF18 in serum, plasma (heparin, EDTA) and cell culture supernatants. Tested Reactivity Hu Tested Application ELISA Specificity There is no detectable cross-reactivity with other relevant proteins. Target Name TNFRSF18 Conjugation HRP Conjugation Note Substrate: TMB and read at 450 nm. Sensitivity 31.25 pg/ml Sample Type Serum, plasma (heparin, EDTA) and cell culture supernatants. Standard Range 62.5 - 4000 pg/ml Sample Volume 100 µl www.arigobio.com 1/3 Precision Intra-Assay CV: 6.3% Inter-Assay CV: 7.0% Alternate Names Tumor necrosis factor receptor superfamily member 18; AITR; CD357; CD antigen CD357; Activation- inducible TNFR family receptor; GITR-D; GITR; Glucocorticoid-induced TNFR-related protein Application Instructions Assay Time ~ 5 hours Properties Form 96 well Storage instruction Store the kit at 2-8°C. -
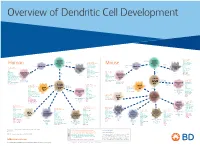
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

BTLA−HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a Cona-Induced Hepatitis Model in Zebrafish
BTLA−HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a ConA-Induced Hepatitis Model in Zebrafish This information is current as Wei Shi, Tong Shao, Jiang-yuan Li, Dong-dong Fan, Ai-fu of September 27, 2021. Lin, Li-xin Xiang and Jian-zhong Shao J Immunol published online 27 September 2019 http://www.jimmunol.org/content/early/2019/09/26/jimmun ol.1900458 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2019/09/26/jimmunol.190045 Material 8.DCSupplemental http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 27, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2019 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published September 27, 2019, doi:10.4049/jimmunol.1900458 The Journal of Immunology BTLA–HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a ConA-Induced Hepatitis Model in Zebrafish Wei Shi,* Tong Shao,* Jiang-yuan Li,* Dong-dong Fan,* Ai-fu Lin,* Li-xin Xiang,* and Jian-zhong Shao*,† The BTLA2HVEM checkpoint axis plays extensive roles in immunomodulation and diseases, including cancer and autoimmune disorders. -

TNFRSF18 (Human) ELISA Kit
TNFRSF18 (Human) ELISA Kit Catalog Number KA1720 96 assays Version: 01 Intended for research use only www.abnova.com I. INTRODUCTION GITR (glucocorticoid induced tumor necrosis factor receptor family related gene) is a type-1 transmembrane protein of 228 amino acids belonging to the TNF and NGF receptor family of proteins. GITR is expressed in normal T-lymphocytes from thymus, spleen. Constitutive expression of a transfected GITR gene induces resistance to apoptosis induced by anti CD3 monoclonal antibodies. The human homolog of GITR is expressed in lymph node and peripheral blood leukocytes. Its expression is up-regulated in human peripheral mononuclear cells mainly after stimulation with antibodies against CD3 and CD28. TNFRSF18 (Human) ELISA Kit is an in vitro enzyme-linked immunosorbent assay for the quantitative measurement of human GITR in cell lysate and tissue lysate. This assay employs an antibody specific for human GITR coated on a 96-well plate. Standards and samples are pipetted into the wells and GITR present in a sample is bound to the wells by the immobilized antibody. The wells are washed and biotinylated anti-human GITR antibody is added. After washing away unbound biotinylated antibody, HRP-conjugated streptavidin is pipetted to the wells. The wells are again washed, a TMB substrate solution is added to the wells and color develops in proportion to the amount of GITR bound. The Stop Solution changes the color from blue to yellow, and the intensity of the color is measured at 450 nm. II. REAGENTS 1. GITR Microplate (Item A): 96 wells (12 strips x 8 wells) coated with anti-human GITR. -

Defective BTLA Functionality Is Rescued by Restoring Lipid Metabolism in Lupus CD4+ T Cells
Defective BTLA functionality is rescued by restoring lipid metabolism in lupus CD4+ T cells Matthieu Sawaf, … , Hélène Dumortier, Fanny Monneaux JCI Insight. 2018;3(13):e99711. https://doi.org/10.1172/jci.insight.99711. Research Article Immunology Coinhibitory receptors play an important role in the prevention of autoimmune diseases, such as systemic lupus erythematosus (SLE), by limiting T cell activation. B and T lymphocyte attenuator (BTLA) is an inhibitory receptor, similar to cytotoxic T lymphocyte–associated protein 4 (CTLA-4) and programmed death 1 (PD1), that negatively regulates the immune response. The role of BTLA in the pathogenesis of autoimmune diseases in humans and, more specifically, in SLE is largely unknown. We investigated BTLA expression on various T cell subsets, and we did not observe significant variations of BTLA expression between lupus patients and healthy controls. However, the enhancement of BTLA expression after activation was significantly lower in SLE patients compared with that in healthy controls. Furthermore, we found an impaired capacity of BTLA to inhibit T cell activation in SLE due to a poor BTLA recruitment to the immunological synapse following T cell stimulation. Finally, we demonstrated that defective BTLA function can be corrected by restoring intracellular trafficking and by normalizing the lipid metabolism in lupus CD4+ T cells. Collectively, our results evidence that the BTLA signaling pathway is altered in SLE T cells and highlight the potential of targeting this pathway for the development of new therapeutic strategies in lupus. Find the latest version: https://jci.me/99711/pdf RESEARCH ARTICLE Defective BTLA functionality is rescued by restoring lipid metabolism in lupus CD4+ T cells Matthieu Sawaf,1 Jean-Daniel Fauny,1 Renaud Felten,1,2 Flora Sagez,1,2 Jacques-Eric Gottenberg,1,2 Hélène Dumortier,1 and Fanny Monneaux1 1CNRS, Institut de Biologie Moléculaire et Cellulaire, Immunologie, Immunopathologie et Chimie Thérapeutique, Strasbourg, France. -

Obeticholic Acid, a Synthetic Bile Acid Agonist of the Farnesoid X Receptor, Attenuates Experimental Autoimmune Encephalomyelitis
Obeticholic acid, a synthetic bile acid agonist of the farnesoid X receptor, attenuates experimental autoimmune encephalomyelitis Peggy P. Hoa and Lawrence Steinmana,1 aDepartment of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford CA 94305 Contributed by Lawrence Steinman, December 22, 2015 (sent for review December 14, 2015; reviewed by Irun R. Cohen and Howard L. Weiner) Bile acids are ligands for the nuclear hormone receptor, farnesoid X (10). Activation of FXR has been reported to inhibit vascular receptor (FXR). The bile acid–FXR interaction regulates bile acid synthe- smooth muscle cell (VSMC) inflammation by down-regulating the sis, transport, and cholesterol metabolism. Recently, bile acid–FXR reg- proinflammatory enzymes inducible nitric oxide synthase and ulation has been reported to play an integral role in both hepatic and cyclooxygenase-2 expression as well as cell migration into VSMCs intestinal inflammation, and in atherosclerosis. In this study, we found (11). This finding suggests that FXR may be a potential target for that FXR knockout mice had more disease severity in experimental the progressive inflammatory disease, atherosclerosis (11, 12). Re- autoimmune encephalomyelitis (EAE), an animal model of multiple cently, 12 nuclear receptors, including FXR, were shown to be + + + + sclerosis (MS). Obeticholic acid (6α-ethyl-chenodeoxycholic acid, expressedonhighlypurifiedCD4 ,CD8 ,CD19 ,andCD14 cells 6-ECDCA), a synthetic FXR agonist, is an orally available drug that is by quantitative real-time PCR, suggesting that FXR may be currently in clinical trials for the treatment of inflammatory diseases coregulatedinhumanimmune cells (13). such as alcoholic hepatitis, nonalcoholic steatohepatitis, and primary Obeticholic acid (6α-ethyl-chenodeoxycholic acid, 6-ECDCA), biliary cirrhosis. -

UNIVERSITY of CALIFORNIA SAN DIEGO Dissection of BTLA
UNIVERSITY OF CALIFORNIA SAN DIEGO Dissection of BTLA Interactomes Using Cell Free Reconstitution A Thesis submitted in partial satisfaction of the requirements for the degree Master of Science in Biology by Zijun Wu Committee in charge: Professor Enfu Hui, Chair Professor Maho Niwa Rosen, Co-Chair Professor Douglass Forbes 2018 Copyright Zijun Wu, 2018 All rights reserved. The Thesis of Zijun Wu is approved and it is acceptable in quality and form for publication electronically: Co-Chair Chair University of California San Diego 2018 iii TABLE OF CONTENTS Signature Page................................................................................................................................iii Table of Contents............................................................................................................................iv List of Abbreviations......................................................................................................................vi List of Figures................................................................................................................................vii Acknowledgements......................................................................................................................viii Abstract of the Thesis.....................................................................................................................ix Introduction......................................................................................................................................1 -

Supplementary Tables
Supplementary Table 1 Lymphocyte activation Activation of immune response Cell adhesion Rank Gene p-value Fold change Rank Gene p-value Fold change Rank Gene p-value Fold change 1 BATF 0.00001 6.31564 1 DUSP6 0.00002 6.39359 1 IL21 0.00014 8.20000 2 MIF 0.00007 2.68925 2 TANK 0.00020 1.69235 2 IL2RA 0.00037 7.70668 3 NFATC2 0.00008 0.40405 3 IRAK1 0.00057 1.63217 3 C1QBP 0.00039 3.79258 4 INPP5D 0.00028 0.31780 4 CHUK 0.00080 1.59486 4 CCL28 0.00010 2.43756 5 TNFSF12 0.00002 0.30456 5 HRAS 0.00004 1.58111 5 BCL10 0.00036 0.62896 6 CASP8 0.00001 0.23611 6 CREB1 0.00037 0.58528 6 IRF1 0.00027 0.62762 7 SMAD3 0.00002 0.23043 7 CD3E 0.00023 0.49823 7 CD46 0.00033 0.52342 8 RORA 0.00001 0.14902 8 ZAP70 0.00068 0.46392 8 SPN 0.00022 0.48806 9 CTSS 0.00024 0.30270 9 JAK2 0.00086 0.45576 10 PLA2G6 0.00002 0.22338 10 TGFB1 0.00006 0.44888 11 TLR3 0.00045 0.13067 11 IL12RB1 0.00021 0.39881 12 ITGA4 0.00098 0.37128 13 ITGAL 0.00013 0.32985 14 CCR2 0.00049 0.29760 15 CD44 0.00097 0.21387 Supplementary Table 1. Differentially expressed genes (p<0.05) in CAR compared to delta CAR T cells. The table displays the p-values and fold changes of genes that are differentially regulated in CAR T cells compared to the truncated CAR (ΔCAR) control T cells collected 24 hours post-stimulation with PSCA. -

Cx3cr1 Mediates the Development of Monocyte-Derived Dendritic Cells During Hepatic Inflammation
CX3CR1 MEDIATES THE DEVELOPMENT OF MONOCYTE-DERIVED DENDRITIC CELLS DURING HEPATIC INFLAMMATION. Supplementary material Supplementary Figure 1: Liver CD45+ myeloid cells were pre-gated for Ly6G negative cells for excluding granulocytes and HDCs subsequently analyzed among the cells that were CD11c+ and had high expression of MHCII. Supplementary Table 1 low/- high + Changes in gene expression between CX3CR1 and CX3CR1 CD11b myeloid hepatic dendritic cells (HDCs) from CCl4-treated mice high Genes up-regulated in CX3CR1 HDCs Gene Fold changes P value Full name App 4,01702 5,89E-05 amyloid beta (A4) precursor protein C1qa 9,75881 1,69E-22 complement component 1, q subcomponent, alpha polypeptide C1qb 9,19882 3,62E-20 complement component 1, q subcomponent, beta polypeptide Ccl12 2,51899 0,011769 chemokine (C-C motif) ligand 12 Ccl2 6,53486 6,37E-11 chemokine (C-C motif) ligand 2 Ccl3 4,99649 5,84E-07 chemokine (C-C motif) ligand 3 Ccl4 4,42552 9,62E-06 chemokine (C-C motif) ligand 4 Ccl6 3,9311 8,46E-05 chemokine (C-C motif) ligand 6 Ccl7 2,60184 0,009272 chemokine (C-C motif) ligand 7 Ccl9 4,17294 3,01E-05 chemokine (C-C motif) ligand 9 Ccr2 3,35195 0,000802 chemokine (C-C motif) receptor 2 Ccr5 3,23358 0,001222 chemokine (C-C motif) receptor 5 Cd14 6,13325 8,61E-10 CD14 antigen Cd36 2,94367 0,003243 CD36 antigen Cd44 4,89958 9,60E-07 CD44 antigen Cd81 6,49623 8,24E-11 CD81 antigen Cd9 3,06253 0,002195 CD9 antigen Cdkn1a 4,65279 3,27E-06 cyclin-dependent kinase inhibitor 1A (P21) Cebpb 6,6083 3,89E-11 CCAAT/enhancer binding protein (C/EBP),