UNIVERSITY of CALIFORNIA SAN DIEGO Dissection of BTLA
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ENSG Gene Encodes Effector TCR Pathway Costimulation Inhibitory/Exhaustion Synapse/Adhesion Chemokines/Receptors
ENSG Gene Encodes Effector TCR pathway Costimulation Inhibitory/exhaustion Synapse/adhesion Chemokines/receptors ENSG00000111537 IFNG IFNg x ENSG00000109471 IL2 IL-2 x ENSG00000232810 TNF TNFa x ENSG00000271503 CCL5 CCL5 x x ENSG00000139187 KLRG1 Klrg1 x ENSG00000117560 FASLG Fas ligand x ENSG00000121858 TNFSF10 TRAIL x ENSG00000134545 KLRC1 Klrc1 / NKG2A x ENSG00000213809 KLRK1 Klrk1 / NKG2D x ENSG00000188389 PDCD1 PD-1 x x ENSG00000117281 CD160 CD160 x x ENSG00000134460 IL2RA IL-2 receptor x subunit alpha ENSG00000110324 IL10RA IL-10 receptor x subunit alpha ENSG00000115604 IL18R1 IL-18 receptor 1 x ENSG00000115607 IL18RAP IL-18 receptor x accessory protein ENSG00000081985 IL12RB2 IL-12 receptor x beta 2 ENSG00000186810 CXCR3 CXCR3 x x ENSG00000005844 ITGAL CD11a x ENSG00000160255 ITGB2 CD18; Integrin x x beta-2 ENSG00000156886 ITGAD CD11d x ENSG00000140678 ITGAX; CD11c x x Integrin alpha-X ENSG00000115232 ITGA4 CD49d; Integrin x x alpha-4 ENSG00000169896 ITGAM CD11b; Integrin x x alpha-M ENSG00000138378 STAT4 Stat4 x ENSG00000115415 STAT1 Stat1 x ENSG00000170581 STAT2 Stat2 x ENSG00000126561 STAT5a Stat5a x ENSG00000162434 JAK1 Jak1 x ENSG00000100453 GZMB Granzyme B x ENSG00000145649 GZMA Granzyme A x ENSG00000180644 PRF1 Perforin 1 x ENSG00000115523 GNLY Granulysin x ENSG00000100450 GZMH Granzyme H x ENSG00000113088 GZMK Granzyme K x ENSG00000057657 PRDM1 Blimp-1 x ENSG00000073861 TBX21 T-bet x ENSG00000115738 ID2 ID2 x ENSG00000176083 ZNF683 Hobit x ENSG00000137265 IRF4 Interferon x regulatory factor 4 ENSG00000140968 IRF8 Interferon -

SLPI and Soluble BTLA As Immunological Markers in Severe Bacterial Infections
SLPI and soluble BTLA as immunological markers in severe bacterial infections To my family Örebro Studies in Medicine 211 ANNA LANGE SLPI and soluble BTLA as immunological markers in severe bacterial infections © Anna Lange, 2020 Title: SLPI and soluble BTLA as immunological markers in severe bacterial infections Publisher: Örebro University 2020 www.oru.se/publikationer Print: Örebro University, Repro 04/2020 ISSN 1652-4063 ISBN 978-91-7529-335-6 Abstract Anna Lange (2020): SLPI and soluble BTLA as immunological markers in severe bacterial infections. Örebro Studies in Medicine 211. Clinical presentation, and outcome of infections are affected by host-, and etiology- (focus of infection and pathogen) related factors. The im- mune response is controlled by a network of regulating pathways. This thesis focuses on Secretory Leukocyte Protease Inhibitor (SLPI), a protease inhibitor with anti-inflammatory properties, and the previously non-studied soluble isoform of B and T lymphocyte attenuator (sBTLA), a membrane-associated regulatory protein. Plasma concentrations of SLPI and sBTLA were assessed in relation to etiology, severity, mortality, and markers of inflammation and immunosuppression, in i) community- acquired pneumonia (CAP) (SLPI), ii) intensive care unit (ICU) treated severe sepsis and septic shock (sBTLA), and iii) dynamically in BSI (SLPI and sBTLA). Main findings were: higher expression of SLPI in pneumonia, com- pared to other sources, higher initial concentrations in Streptococcus pneumoniae, and Staphylococcus aureus BSI, compared to Escherichia coli BSI, and higher SLPI concentrations in sepsis compared to non-septic BSI. Interestingly, men with pneumonia had higher plasma levels of SLPI, both in CAP and BSI. Likewise, sBTLA was associated with severity, but preferentially at higher organ failure scores. -
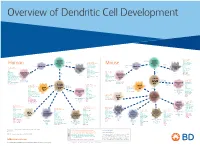
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

BTLA−HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a Cona-Induced Hepatitis Model in Zebrafish
BTLA−HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a ConA-Induced Hepatitis Model in Zebrafish This information is current as Wei Shi, Tong Shao, Jiang-yuan Li, Dong-dong Fan, Ai-fu of September 27, 2021. Lin, Li-xin Xiang and Jian-zhong Shao J Immunol published online 27 September 2019 http://www.jimmunol.org/content/early/2019/09/26/jimmun ol.1900458 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2019/09/26/jimmunol.190045 Material 8.DCSupplemental http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 27, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2019 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published September 27, 2019, doi:10.4049/jimmunol.1900458 The Journal of Immunology BTLA–HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a ConA-Induced Hepatitis Model in Zebrafish Wei Shi,* Tong Shao,* Jiang-yuan Li,* Dong-dong Fan,* Ai-fu Lin,* Li-xin Xiang,* and Jian-zhong Shao*,† The BTLA2HVEM checkpoint axis plays extensive roles in immunomodulation and diseases, including cancer and autoimmune disorders. -

Defective BTLA Functionality Is Rescued by Restoring Lipid Metabolism in Lupus CD4+ T Cells
Defective BTLA functionality is rescued by restoring lipid metabolism in lupus CD4+ T cells Matthieu Sawaf, … , Hélène Dumortier, Fanny Monneaux JCI Insight. 2018;3(13):e99711. https://doi.org/10.1172/jci.insight.99711. Research Article Immunology Coinhibitory receptors play an important role in the prevention of autoimmune diseases, such as systemic lupus erythematosus (SLE), by limiting T cell activation. B and T lymphocyte attenuator (BTLA) is an inhibitory receptor, similar to cytotoxic T lymphocyte–associated protein 4 (CTLA-4) and programmed death 1 (PD1), that negatively regulates the immune response. The role of BTLA in the pathogenesis of autoimmune diseases in humans and, more specifically, in SLE is largely unknown. We investigated BTLA expression on various T cell subsets, and we did not observe significant variations of BTLA expression between lupus patients and healthy controls. However, the enhancement of BTLA expression after activation was significantly lower in SLE patients compared with that in healthy controls. Furthermore, we found an impaired capacity of BTLA to inhibit T cell activation in SLE due to a poor BTLA recruitment to the immunological synapse following T cell stimulation. Finally, we demonstrated that defective BTLA function can be corrected by restoring intracellular trafficking and by normalizing the lipid metabolism in lupus CD4+ T cells. Collectively, our results evidence that the BTLA signaling pathway is altered in SLE T cells and highlight the potential of targeting this pathway for the development of new therapeutic strategies in lupus. Find the latest version: https://jci.me/99711/pdf RESEARCH ARTICLE Defective BTLA functionality is rescued by restoring lipid metabolism in lupus CD4+ T cells Matthieu Sawaf,1 Jean-Daniel Fauny,1 Renaud Felten,1,2 Flora Sagez,1,2 Jacques-Eric Gottenberg,1,2 Hélène Dumortier,1 and Fanny Monneaux1 1CNRS, Institut de Biologie Moléculaire et Cellulaire, Immunologie, Immunopathologie et Chimie Thérapeutique, Strasbourg, France. -

Obeticholic Acid, a Synthetic Bile Acid Agonist of the Farnesoid X Receptor, Attenuates Experimental Autoimmune Encephalomyelitis
Obeticholic acid, a synthetic bile acid agonist of the farnesoid X receptor, attenuates experimental autoimmune encephalomyelitis Peggy P. Hoa and Lawrence Steinmana,1 aDepartment of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford CA 94305 Contributed by Lawrence Steinman, December 22, 2015 (sent for review December 14, 2015; reviewed by Irun R. Cohen and Howard L. Weiner) Bile acids are ligands for the nuclear hormone receptor, farnesoid X (10). Activation of FXR has been reported to inhibit vascular receptor (FXR). The bile acid–FXR interaction regulates bile acid synthe- smooth muscle cell (VSMC) inflammation by down-regulating the sis, transport, and cholesterol metabolism. Recently, bile acid–FXR reg- proinflammatory enzymes inducible nitric oxide synthase and ulation has been reported to play an integral role in both hepatic and cyclooxygenase-2 expression as well as cell migration into VSMCs intestinal inflammation, and in atherosclerosis. In this study, we found (11). This finding suggests that FXR may be a potential target for that FXR knockout mice had more disease severity in experimental the progressive inflammatory disease, atherosclerosis (11, 12). Re- autoimmune encephalomyelitis (EAE), an animal model of multiple cently, 12 nuclear receptors, including FXR, were shown to be + + + + sclerosis (MS). Obeticholic acid (6α-ethyl-chenodeoxycholic acid, expressedonhighlypurifiedCD4 ,CD8 ,CD19 ,andCD14 cells 6-ECDCA), a synthetic FXR agonist, is an orally available drug that is by quantitative real-time PCR, suggesting that FXR may be currently in clinical trials for the treatment of inflammatory diseases coregulatedinhumanimmune cells (13). such as alcoholic hepatitis, nonalcoholic steatohepatitis, and primary Obeticholic acid (6α-ethyl-chenodeoxycholic acid, 6-ECDCA), biliary cirrhosis. -

Cx3cr1 Mediates the Development of Monocyte-Derived Dendritic Cells During Hepatic Inflammation
CX3CR1 MEDIATES THE DEVELOPMENT OF MONOCYTE-DERIVED DENDRITIC CELLS DURING HEPATIC INFLAMMATION. Supplementary material Supplementary Figure 1: Liver CD45+ myeloid cells were pre-gated for Ly6G negative cells for excluding granulocytes and HDCs subsequently analyzed among the cells that were CD11c+ and had high expression of MHCII. Supplementary Table 1 low/- high + Changes in gene expression between CX3CR1 and CX3CR1 CD11b myeloid hepatic dendritic cells (HDCs) from CCl4-treated mice high Genes up-regulated in CX3CR1 HDCs Gene Fold changes P value Full name App 4,01702 5,89E-05 amyloid beta (A4) precursor protein C1qa 9,75881 1,69E-22 complement component 1, q subcomponent, alpha polypeptide C1qb 9,19882 3,62E-20 complement component 1, q subcomponent, beta polypeptide Ccl12 2,51899 0,011769 chemokine (C-C motif) ligand 12 Ccl2 6,53486 6,37E-11 chemokine (C-C motif) ligand 2 Ccl3 4,99649 5,84E-07 chemokine (C-C motif) ligand 3 Ccl4 4,42552 9,62E-06 chemokine (C-C motif) ligand 4 Ccl6 3,9311 8,46E-05 chemokine (C-C motif) ligand 6 Ccl7 2,60184 0,009272 chemokine (C-C motif) ligand 7 Ccl9 4,17294 3,01E-05 chemokine (C-C motif) ligand 9 Ccr2 3,35195 0,000802 chemokine (C-C motif) receptor 2 Ccr5 3,23358 0,001222 chemokine (C-C motif) receptor 5 Cd14 6,13325 8,61E-10 CD14 antigen Cd36 2,94367 0,003243 CD36 antigen Cd44 4,89958 9,60E-07 CD44 antigen Cd81 6,49623 8,24E-11 CD81 antigen Cd9 3,06253 0,002195 CD9 antigen Cdkn1a 4,65279 3,27E-06 cyclin-dependent kinase inhibitor 1A (P21) Cebpb 6,6083 3,89E-11 CCAAT/enhancer binding protein (C/EBP), -

CD160 and BTLA: Lights out for CD4+ T Cells
NEWS AND VIEWS Foxp3 expression. The fact that such binding β-dependent induction does not yield a very 3. Marie, J.C., Letterio, J.J., Gavin, M. & Rudensky, A.Y. + J. Exp. Med. 201, 1061–1067 (2005). causes H4 acetylation and that such acetyla- stable phenotype of Foxp3 Treg cells and thus 4. Huber, S. et al. J. Immunol. 173, 6526–6531 + tion is only found in Foxp3 cells constitutes other factors may contribute in vivo as well. At (2004). a good argument for this claim. However, this point, the results of Tone et al.2 raise the 5. Kretschmer, K. et al. Nat. Immunol. 6, 1219–1227 (2005). these results do not rule out that there is a question of whether there are Smad3-inde- 6. von Boehmer, H. J. Exp. Med. 204, 1737–1739 Smad3-independent and therefore TGF-β pendent pathways of Foxp3 induction, which (2007). ligand–independent pathway of Foxp3 induc- presumably can be tested by a ‘knock-in’ Foxp3 7. Burchill, M.A., Yang, J., Vogtenhuber, C., Blazar, B.R. & Farrar, M.A. J. Immunol. 178, 280–290 (2007). tion. Despite this cautionary note, the authors allele with a mutated Smad3 binding site. 8. Kim, H.P. & Leonard, W.J. J. Exp. Med. 204, 1543– seem to have established a pathway of TGF-β- 1551 (2007). 9. Chen, W. et al. J. Exp. Med. 198, 1875–1886 dependent induction of Foxp3 expression. It has 1. Brunkow, M.E. et al. Nat. Genet. 27, 68–73 (2001). (2003). been known for some time that in vitro TGF- 2. -

Blockade of HVEM for Prostate Cancer Immunotherapy in Humanized Mice
cancers Article Blockade of HVEM for Prostate Cancer Immunotherapy in Humanized Mice Nicolas Aubert 1,† , Simon Brunel 1,†, Daniel Olive 2 and Gilles Marodon 1,* 1 Centre d’Immunologie et Maladies Infectieuses-Paris, CIMI-PARIS, Sorbonne Université, INSERM, CNRS, 75013 Paris, France; [email protected] (N.A.); [email protected] (S.B.) 2 Institut Paoli-Calmettes, Aix-Marseille Université, INSERM, CNRS, CRCM, Tumor Immunity Team, IBISA Immunomonitoring Platform, 13009 Marseille, France; [email protected] * Correspondence: [email protected] † Equal contributions. Simple Summary: Current immune checkpoint inhibitors have shown limitations for immunother- apy of prostate cancer. Thus, it is crucial to investigate other immune checkpoints to prevent disease progression in patients with prostate cancer. Here, we first show that the HVEM/BTLA immune checkpoint is associated with disease progression in patients. We then show that immunotherapy aimed at targeting HVEM reduced tumor growth twofold in vivo in a humanized mouse model of the pathology. The mode of action of the therapy was dependent on CD8+ T cells and is associated with improved T cell activation and reduced exhaustion. Finally, we demonstrated that HVEM expressed by the tumor negatively regulated the anti-tumor immune response. Our results indicate that targeting HVEM might be an attractive option for patients with prostate cancer. Abstract: The herpes virus entry mediator (HVEM) delivers a negative signal to T cells mainly through the B and T lymphocyte attenuator (BTLA) molecule. Thus, HVEM/BTLA may represent a Citation: Aubert, N.; Brunel, S.; novel immune checkpoint during an anti-tumor immune response. However, a formal demonstration Olive, D.; Marodon, G. -

Inhibitory Receptor Agonists: the Future of Autoimmune Disease Therapeutics?
Available online at www.sciencedirect.com ScienceDirect Inhibitory receptor agonists: the future of autoimmune disease therapeutics? 1,2 1 Stephanie Grebinoski and Dario AA Vignali + Central and peripheral tolerance both contribute to protection Tregs, implying that >30% of CD4 effectors could be against autoimmunity. The pathogenesis of autoimmunity, autoreactive [2 ]. Critical deficits or limitations in toler- however, can result from critical deficits or limitations in ance mechanisms, rather than complete failure, can con- peripheral and/or central tolerance mechanisms, presenting an tribute to autoimmunity (Figure 1). Evidence suggests opportunity for therapeutic intervention. Recent advances that the majority of these deficits affect peripheral toler- highlight the substantial impact of inhibitory receptors (IRs), ance, presenting an opportunity for therapeutic interven- which mediate peripheral tolerance, in autoimmunity. Deletion tion [1]. and blockade studies in mice, IR disruption in humans, and correlation with positive disease outcomes all highlight Recent advances in cancer immunotherapy indicate that IR potential clinical benefits of enhancing IR signaling modulation affects T cell function and disease outcome, (agonism) — specifically CTLA4, PD1, LAG3, TIM3 and which merited the 2018 Nobel Prize in Medicine [3]. IRs TIGIT — to treat autoimmune disease. Although critical mediateperipheral tolerance byantagonizing T cell receptor questions remain, IR agonists represent an unappreciated and (TCR) signaling and/or signal propagation in autoreactive T untapped opportunity for the treatment of autoimmune and cells, resulting in a dysfunctional state [4]. Evolutionarily, inflammatory diseases. IRs are upregulated in response TCR signaling to dampen the immune response post infection or upon recognition of Addresses self-antigen(Figure1). Under chronic conditions, IR expres- 1 Department of Immunology, University of Pittsburgh School of sion is heightened and T cell effector function is inhibited Medicine, 200 Lothrop Street, Pittsburgh, PA 15261, USA 2 [5]. -

(BTLA) in CD8 TUMOR INFILTRATING LYMPHOCYTES Krit Ritthipichai
The Texas Medical Center Library DigitalCommons@TMC The University of Texas MD Anderson Cancer Center UTHealth Graduate School of The University of Texas MD Anderson Cancer Biomedical Sciences Dissertations and Theses Center UTHealth Graduate School of (Open Access) Biomedical Sciences 8-2016 MECHANISTIC AND FUNCTIONAL CHARACTERIZATION OF B AND T LYMPHOCYTE ATTENUATOR (BTLA) IN CD8 TUMOR INFILTRATING LYMPHOCYTES krit ritthipichai Follow this and additional works at: https://digitalcommons.library.tmc.edu/utgsbs_dissertations Part of the Medicine and Health Sciences Commons Recommended Citation ritthipichai, krit, "MECHANISTIC AND FUNCTIONAL CHARACTERIZATION OF B AND T LYMPHOCYTE ATTENUATOR (BTLA) IN CD8 TUMOR INFILTRATING LYMPHOCYTES" (2016). The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access). 685. https://digitalcommons.library.tmc.edu/utgsbs_dissertations/685 This Dissertation (PhD) is brought to you for free and open access by the The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences at DigitalCommons@TMC. It has been accepted for inclusion in The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access) by an authorized administrator of DigitalCommons@TMC. For more information, please contact [email protected]. MECHANISTIC AND FUNCTIONAL CHARACTERIZATION OF B AND T LYMPHOCYTE ATTENUATOR (BTLA) IN CD8 TUMOR INFILTRATING -

Anti–IL-7 Receptor-Α Reverses Established Type 1 Diabetes In
Correction IMMUNOLOGY Correction for “Anti–IL-7 receptor-α reverses established type 1 diabetes in nonobese diabetic mice by modulating effector T-cell function,” by Li-Fen Lee, Kathryn Logronio, Guang Huan Tu, Wenwu Zhai, Irene Ni, Li Mei, Jeanette Dilley, Jessica Yu, Arvind Rajpal, Colleen Brown, Charles Appah, Sherman Mi- chael Chin, Bora Han, Timothy Affolter, and John C. Lin, which appeared in issue 31, July 31, 2012, of Proc Natl Acad Sci USA (109:12674–12679; first published June 25, 2012; 10.1073/ pnas.1203795109). The authors note that their conflict of interest statement was omitted during publication. The authors declare that all the authors are full-time employees of Pfizer Inc. Pfizer Inc. filed a patent application, with L.F.-L., J.C.L., and W.Z. as co- inventors: US Application Serial No. 13/033,491, entitled “Antagonist Anti-IL-7 Receptor Antibodies and Methods.” www.pnas.org/cgi/doi/10.1073/pnas.1214896109 CORRECTION www.pnas.org PNAS | October 2, 2012 | vol. 109 | no. 40 | 16393 Downloaded by guest on September 23, 2021 Anti–IL-7 receptor-α reverses established type 1 diabetes in nonobese diabetic mice by modulating effector T-cell function Li-Fen Leea,1, Kathryn Logronioa, Guang Huan Tua, Wenwu Zhaia, Irene Nia, Li Meia, Jeanette Dilleya, Jessica Yua, Arvind Rajpala, Colleen Browna, Charles Appaha, Sherman Michael Chin a, Bora Hanb,TimothyAffolterb, and John C. Lina,1 aRinat, Pfizer Inc., South San Francisco, CA 94080; and bDrug Safety R and D, Pfizer Inc., La Jolla, CA 92121 Edited by Lewis L. Lanier, University of California, San Francisco, CA, and approved May 28, 2012 (received for review March 7, 2012) Genetic variation in the IL-7 receptor-α (IL-7R) gene is associated influence the development of the IFN-γ–expressing CD8+ with susceptibility to human type 1 diabetes (T1D).