(BTLA) in CD8 TUMOR INFILTRATING LYMPHOCYTES Krit Ritthipichai
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ENSG Gene Encodes Effector TCR Pathway Costimulation Inhibitory/Exhaustion Synapse/Adhesion Chemokines/Receptors
ENSG Gene Encodes Effector TCR pathway Costimulation Inhibitory/exhaustion Synapse/adhesion Chemokines/receptors ENSG00000111537 IFNG IFNg x ENSG00000109471 IL2 IL-2 x ENSG00000232810 TNF TNFa x ENSG00000271503 CCL5 CCL5 x x ENSG00000139187 KLRG1 Klrg1 x ENSG00000117560 FASLG Fas ligand x ENSG00000121858 TNFSF10 TRAIL x ENSG00000134545 KLRC1 Klrc1 / NKG2A x ENSG00000213809 KLRK1 Klrk1 / NKG2D x ENSG00000188389 PDCD1 PD-1 x x ENSG00000117281 CD160 CD160 x x ENSG00000134460 IL2RA IL-2 receptor x subunit alpha ENSG00000110324 IL10RA IL-10 receptor x subunit alpha ENSG00000115604 IL18R1 IL-18 receptor 1 x ENSG00000115607 IL18RAP IL-18 receptor x accessory protein ENSG00000081985 IL12RB2 IL-12 receptor x beta 2 ENSG00000186810 CXCR3 CXCR3 x x ENSG00000005844 ITGAL CD11a x ENSG00000160255 ITGB2 CD18; Integrin x x beta-2 ENSG00000156886 ITGAD CD11d x ENSG00000140678 ITGAX; CD11c x x Integrin alpha-X ENSG00000115232 ITGA4 CD49d; Integrin x x alpha-4 ENSG00000169896 ITGAM CD11b; Integrin x x alpha-M ENSG00000138378 STAT4 Stat4 x ENSG00000115415 STAT1 Stat1 x ENSG00000170581 STAT2 Stat2 x ENSG00000126561 STAT5a Stat5a x ENSG00000162434 JAK1 Jak1 x ENSG00000100453 GZMB Granzyme B x ENSG00000145649 GZMA Granzyme A x ENSG00000180644 PRF1 Perforin 1 x ENSG00000115523 GNLY Granulysin x ENSG00000100450 GZMH Granzyme H x ENSG00000113088 GZMK Granzyme K x ENSG00000057657 PRDM1 Blimp-1 x ENSG00000073861 TBX21 T-bet x ENSG00000115738 ID2 ID2 x ENSG00000176083 ZNF683 Hobit x ENSG00000137265 IRF4 Interferon x regulatory factor 4 ENSG00000140968 IRF8 Interferon -

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Cd8a (Ly-2) Microbeads Mouse
CD8a (Ly-2) MicroBeads mouse Order no. 130-049-401 Contents 1.2 Background information 1. Description Mouse CD8a (Ly-2) MicroBeads were developed for positive + 1.1 Principle of the MACS® Separation selection or depletion of mouse CD8a T cells from single-cell suspensions of lymphoid and non-lymphoid tissues or from 1.2 Background information peripheral blood. The CD8a antigen is expressed on most 1.3 Applications thymocytes, almost all cytotoxic T cells and on subpopulations of dendritic cells. CD8a functions as an accessory molecule in 1.4 Reagent and instrument requirements the recognition of MHC class I/peptide complexes by the TCR 2. Protocol heterodimer on cytotoxic CD8a+ T cells. 2.1 Sample preparation 1.3 Applications 2.2 Magnetic labeling + 2.3 Magnetic separation ● Positive selection or depletion of CD8a T cells from lymphoid organs, non-lymphoid tissue, peripheral blood, or in vitro 2.4 Cell separation with the autoMACS® Pro Separator cultured cells. 3. Example of a separation using the CD8a (Ly-2) MicroBeads + ● Isolation of purified CD8 cells for in vitro and in vivo studies 1,2 4. References on protective immune responses against parasites or allergens3, and for adoptive transfer into immunodeficient4,5 and virus infected mice6. + Warnings ● Isolation of highly pure CD8 T cells from CNS of MHV infected 7 Reagents contain sodium azide. Under acidic conditions sodium mice for evaluation of their chemokine expression pattern. azide yields hydrazoic acid, which is extremely toxic. Azide compounds should be diluted with running water before discarding. 1.4 Reagent and instrument requirements These precautions are recommended to avoid deposits in plumbing ● Buffer: Prepare a solution containing phosphate-buffered where explosive conditions may develop. -

SLPI and Soluble BTLA As Immunological Markers in Severe Bacterial Infections
SLPI and soluble BTLA as immunological markers in severe bacterial infections To my family Örebro Studies in Medicine 211 ANNA LANGE SLPI and soluble BTLA as immunological markers in severe bacterial infections © Anna Lange, 2020 Title: SLPI and soluble BTLA as immunological markers in severe bacterial infections Publisher: Örebro University 2020 www.oru.se/publikationer Print: Örebro University, Repro 04/2020 ISSN 1652-4063 ISBN 978-91-7529-335-6 Abstract Anna Lange (2020): SLPI and soluble BTLA as immunological markers in severe bacterial infections. Örebro Studies in Medicine 211. Clinical presentation, and outcome of infections are affected by host-, and etiology- (focus of infection and pathogen) related factors. The im- mune response is controlled by a network of regulating pathways. This thesis focuses on Secretory Leukocyte Protease Inhibitor (SLPI), a protease inhibitor with anti-inflammatory properties, and the previously non-studied soluble isoform of B and T lymphocyte attenuator (sBTLA), a membrane-associated regulatory protein. Plasma concentrations of SLPI and sBTLA were assessed in relation to etiology, severity, mortality, and markers of inflammation and immunosuppression, in i) community- acquired pneumonia (CAP) (SLPI), ii) intensive care unit (ICU) treated severe sepsis and septic shock (sBTLA), and iii) dynamically in BSI (SLPI and sBTLA). Main findings were: higher expression of SLPI in pneumonia, com- pared to other sources, higher initial concentrations in Streptococcus pneumoniae, and Staphylococcus aureus BSI, compared to Escherichia coli BSI, and higher SLPI concentrations in sepsis compared to non-septic BSI. Interestingly, men with pneumonia had higher plasma levels of SLPI, both in CAP and BSI. Likewise, sBTLA was associated with severity, but preferentially at higher organ failure scores. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -
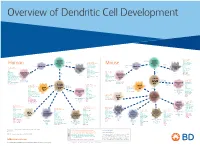
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Changes in Peripheral and Local Tumor Immunity After Neoadjuvant Chemotherapy Reshape Clinical Outcomes in Patients with Breast Cancer Margaret L
Published OnlineFirst August 21, 2020; DOI: 10.1158/1078-0432.CCR-19-3685 CLINICAL CANCER RESEARCH | TRANSLATIONAL CANCER MECHANISMS AND THERAPY Changes in Peripheral and Local Tumor Immunity after Neoadjuvant Chemotherapy Reshape Clinical Outcomes in Patients with Breast Cancer Margaret L. Axelrod1, Mellissa J. Nixon1, Paula I. Gonzalez-Ericsson2, Riley E. Bergman1, Mark A. Pilkinton3, Wyatt J. McDonnell3, Violeta Sanchez1,2, Susan R. Opalenik1, Sherene Loi4, Jing Zhou5, Sean Mackay5, Brent N. Rexer1, Vandana G. Abramson1, Valerie M. Jansen1, Simon Mallal3, Joshua Donaldson1, Sara M. Tolaney6, Ian E. Krop6, Ana C. Garrido-Castro6, Jonathan D. Marotti7,8, Kevin Shee9, Todd. W. Miller8,9, Melinda E. Sanders2,10, Ingrid A. Mayer1,2, Roberto Salgado4,11, and Justin M. Balko1,2 ABSTRACT ◥ Purpose: The recent approval of anti-programmed death-ligand Results: In non-TNBC, no change in expression of any single 1 immunotherapy in combination with nab-paclitaxel for meta- gene was associated with RFS or OS, while in TNBC upregulation of static triple-negative breast cancer (TNBC) highlights the need to multiple immune-related genes and gene sets were associated with understand the role of chemotherapy in modulating the tumor improved long-term outcome. High cytotoxic T-cell signatures immune microenvironment (TIME). present in the peripheral blood of patients with breast cancer at Experimental Design: We examined immune-related gene surgery were associated with persistent disease and recurrence, expression patterns before and after neoadjuvant chemotherapy suggesting active antitumor immunity that may indicate ongoing (NAC) in a series of 83 breast tumors, including 44 TNBCs, from disease burden. patients with residual disease (RD). -

BTLA−HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a Cona-Induced Hepatitis Model in Zebrafish
BTLA−HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a ConA-Induced Hepatitis Model in Zebrafish This information is current as Wei Shi, Tong Shao, Jiang-yuan Li, Dong-dong Fan, Ai-fu of September 27, 2021. Lin, Li-xin Xiang and Jian-zhong Shao J Immunol published online 27 September 2019 http://www.jimmunol.org/content/early/2019/09/26/jimmun ol.1900458 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2019/09/26/jimmunol.190045 Material 8.DCSupplemental http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 27, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2019 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published September 27, 2019, doi:10.4049/jimmunol.1900458 The Journal of Immunology BTLA–HVEM Checkpoint Axis Regulates Hepatic Homeostasis and Inflammation in a ConA-Induced Hepatitis Model in Zebrafish Wei Shi,* Tong Shao,* Jiang-yuan Li,* Dong-dong Fan,* Ai-fu Lin,* Li-xin Xiang,* and Jian-zhong Shao*,† The BTLA2HVEM checkpoint axis plays extensive roles in immunomodulation and diseases, including cancer and autoimmune disorders. -

Roles of Activating Functions 1 and 2 of Estrogen Receptor Α in Lymphopoiesis
236 2 Journal of A Andersson et al. ERαAF-1 and ERαAF-2 in 236:2 99–109 Endocrinology ER-mediated immunomodulation RESEARCH Roles of activating functions 1 and 2 of estrogen receptor α in lymphopoiesis Annica Andersson1, Anna E Törnqvist2, Sofia Moverare-Skrtic2, Angelina I Bernardi1, Helen H Farman2, Pierre Chambon3, Cecilia Engdahl1,2, Marie K Lagerquist2, Sara H Windahl2, Hans Carlsten1, Claes Ohlsson2 and Ulrika Islander1 1Centre for Bone and Arthritis Research, Department of Rheumatology and Inflammation Research, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden 2Centre for Bone and Arthritis Research, Department of Internal Medicine and Clinical Nutrition, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden 3Institut de Génétique et de Biologie Moléculaire et Cellulaire, Centre National de la Recherche Scientifique, National de la Sante et de la Recherche Medicale, ULP, Collège de France, Illkirch-Strasbourg, France Correspondence should be addressed to A Andersson: [email protected] Abstract Apart from the role of sex steroids in reproduction, sex steroids are also important Key Words regulators of the immune system. 17β-estradiol (E2) represses T and B cell development, f lymphopoiesis but augments B cell function, possibly explaining the different nature of immune f estrogen receptor alpha responses in men and women. Both E2 and selective estrogen receptors modulators f estradiol (SERM) act via estrogen receptors (ER). Activating functions (AF)-1 and 2 of the ER f selective estrogen bind to coregulators and thus influence target gene transcription and subsequent receptor modulators cellular response to ER activation. The importance of ERαAF-1 and AF-2 in the immunomodulatory effects of E2/SERM has previously not been reported. -

Defective BTLA Functionality Is Rescued by Restoring Lipid Metabolism in Lupus CD4+ T Cells
Defective BTLA functionality is rescued by restoring lipid metabolism in lupus CD4+ T cells Matthieu Sawaf, … , Hélène Dumortier, Fanny Monneaux JCI Insight. 2018;3(13):e99711. https://doi.org/10.1172/jci.insight.99711. Research Article Immunology Coinhibitory receptors play an important role in the prevention of autoimmune diseases, such as systemic lupus erythematosus (SLE), by limiting T cell activation. B and T lymphocyte attenuator (BTLA) is an inhibitory receptor, similar to cytotoxic T lymphocyte–associated protein 4 (CTLA-4) and programmed death 1 (PD1), that negatively regulates the immune response. The role of BTLA in the pathogenesis of autoimmune diseases in humans and, more specifically, in SLE is largely unknown. We investigated BTLA expression on various T cell subsets, and we did not observe significant variations of BTLA expression between lupus patients and healthy controls. However, the enhancement of BTLA expression after activation was significantly lower in SLE patients compared with that in healthy controls. Furthermore, we found an impaired capacity of BTLA to inhibit T cell activation in SLE due to a poor BTLA recruitment to the immunological synapse following T cell stimulation. Finally, we demonstrated that defective BTLA function can be corrected by restoring intracellular trafficking and by normalizing the lipid metabolism in lupus CD4+ T cells. Collectively, our results evidence that the BTLA signaling pathway is altered in SLE T cells and highlight the potential of targeting this pathway for the development of new therapeutic strategies in lupus. Find the latest version: https://jci.me/99711/pdf RESEARCH ARTICLE Defective BTLA functionality is rescued by restoring lipid metabolism in lupus CD4+ T cells Matthieu Sawaf,1 Jean-Daniel Fauny,1 Renaud Felten,1,2 Flora Sagez,1,2 Jacques-Eric Gottenberg,1,2 Hélène Dumortier,1 and Fanny Monneaux1 1CNRS, Institut de Biologie Moléculaire et Cellulaire, Immunologie, Immunopathologie et Chimie Thérapeutique, Strasbourg, France. -

Obeticholic Acid, a Synthetic Bile Acid Agonist of the Farnesoid X Receptor, Attenuates Experimental Autoimmune Encephalomyelitis
Obeticholic acid, a synthetic bile acid agonist of the farnesoid X receptor, attenuates experimental autoimmune encephalomyelitis Peggy P. Hoa and Lawrence Steinmana,1 aDepartment of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford CA 94305 Contributed by Lawrence Steinman, December 22, 2015 (sent for review December 14, 2015; reviewed by Irun R. Cohen and Howard L. Weiner) Bile acids are ligands for the nuclear hormone receptor, farnesoid X (10). Activation of FXR has been reported to inhibit vascular receptor (FXR). The bile acid–FXR interaction regulates bile acid synthe- smooth muscle cell (VSMC) inflammation by down-regulating the sis, transport, and cholesterol metabolism. Recently, bile acid–FXR reg- proinflammatory enzymes inducible nitric oxide synthase and ulation has been reported to play an integral role in both hepatic and cyclooxygenase-2 expression as well as cell migration into VSMCs intestinal inflammation, and in atherosclerosis. In this study, we found (11). This finding suggests that FXR may be a potential target for that FXR knockout mice had more disease severity in experimental the progressive inflammatory disease, atherosclerosis (11, 12). Re- autoimmune encephalomyelitis (EAE), an animal model of multiple cently, 12 nuclear receptors, including FXR, were shown to be + + + + sclerosis (MS). Obeticholic acid (6α-ethyl-chenodeoxycholic acid, expressedonhighlypurifiedCD4 ,CD8 ,CD19 ,andCD14 cells 6-ECDCA), a synthetic FXR agonist, is an orally available drug that is by quantitative real-time PCR, suggesting that FXR may be currently in clinical trials for the treatment of inflammatory diseases coregulatedinhumanimmune cells (13). such as alcoholic hepatitis, nonalcoholic steatohepatitis, and primary Obeticholic acid (6α-ethyl-chenodeoxycholic acid, 6-ECDCA), biliary cirrhosis.