Detection of Immune Checkpoint Receptors &Ndash
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ENSG Gene Encodes Effector TCR Pathway Costimulation Inhibitory/Exhaustion Synapse/Adhesion Chemokines/Receptors
ENSG Gene Encodes Effector TCR pathway Costimulation Inhibitory/exhaustion Synapse/adhesion Chemokines/receptors ENSG00000111537 IFNG IFNg x ENSG00000109471 IL2 IL-2 x ENSG00000232810 TNF TNFa x ENSG00000271503 CCL5 CCL5 x x ENSG00000139187 KLRG1 Klrg1 x ENSG00000117560 FASLG Fas ligand x ENSG00000121858 TNFSF10 TRAIL x ENSG00000134545 KLRC1 Klrc1 / NKG2A x ENSG00000213809 KLRK1 Klrk1 / NKG2D x ENSG00000188389 PDCD1 PD-1 x x ENSG00000117281 CD160 CD160 x x ENSG00000134460 IL2RA IL-2 receptor x subunit alpha ENSG00000110324 IL10RA IL-10 receptor x subunit alpha ENSG00000115604 IL18R1 IL-18 receptor 1 x ENSG00000115607 IL18RAP IL-18 receptor x accessory protein ENSG00000081985 IL12RB2 IL-12 receptor x beta 2 ENSG00000186810 CXCR3 CXCR3 x x ENSG00000005844 ITGAL CD11a x ENSG00000160255 ITGB2 CD18; Integrin x x beta-2 ENSG00000156886 ITGAD CD11d x ENSG00000140678 ITGAX; CD11c x x Integrin alpha-X ENSG00000115232 ITGA4 CD49d; Integrin x x alpha-4 ENSG00000169896 ITGAM CD11b; Integrin x x alpha-M ENSG00000138378 STAT4 Stat4 x ENSG00000115415 STAT1 Stat1 x ENSG00000170581 STAT2 Stat2 x ENSG00000126561 STAT5a Stat5a x ENSG00000162434 JAK1 Jak1 x ENSG00000100453 GZMB Granzyme B x ENSG00000145649 GZMA Granzyme A x ENSG00000180644 PRF1 Perforin 1 x ENSG00000115523 GNLY Granulysin x ENSG00000100450 GZMH Granzyme H x ENSG00000113088 GZMK Granzyme K x ENSG00000057657 PRDM1 Blimp-1 x ENSG00000073861 TBX21 T-bet x ENSG00000115738 ID2 ID2 x ENSG00000176083 ZNF683 Hobit x ENSG00000137265 IRF4 Interferon x regulatory factor 4 ENSG00000140968 IRF8 Interferon -

IL-15 Stimulation with TIGIT Blockade Reverses CD155-Mediated NK Cell
Author Manuscript Published OnlineFirst on June 26, 2020; DOI: 10.1158/1078-0432.CCR-20-0575 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. IL-15 stimulation with TIGIT blockade reverses CD155-mediated NK cell dysfunction in melanoma Joe-Marc Chauvin1, Mignane Ka1, Ornella Pagliano1, Carmine Menna1, Quanquan Ding1, Richelle DeBlasio1, Cindy Sanders1, Jiajie Hou2, Xian-Yang Li3, Soldano Ferrone4, Diwakar Davar1, John M. Kirkwood1, Robert Johnston5, Alan J. Korman5, Mark J. Smyth3, and Hassane M. Zarour1,6. 1Department of Medicine and Division of Hematology/Oncology, University of Pittsburgh, School of Medicine, Pittsburgh, PA 15213, USA. 2Department of Liver Surgery, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200127, China. 3Immunology in Cancer and Infection Laboratory, QIMR Berghofer Medical Research Institute, Queensland, Australia. 4Department of Surgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114, USA. 5Biologics Discovery California, Bristol-Myers Squibb, Redwood City, CA 94063, USA. 6Department of Immunology, University of Pittsburgh, School of Medicine, Pittsburgh, PA 15213, USA. Running title: IL-15 and TIGIT blockade reverse NK dysfunction in melanoma Keywords: Melanoma, Immunotherapy, TIGIT, IL-15, NK cells This work was supported by NIH/NCI grants R01CA228181 and R01CA222203 (to HMZ), a research grant by Bristol-Myers Squibb BMS (to HMZ), a cancer vaccine collaborative clinical strategy team grant (to HMZ), and NCI grant P50CA121973 (to 1 Downloaded from clincancerres.aacrjournals.org on September 25, 2021. © 2020 American Association for Cancer Research. Author Manuscript Published OnlineFirst on June 26, 2020; DOI: 10.1158/1078-0432.CCR-20-0575 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -

Selectins in Cancer Immunity
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2018 Selectins in cancer immunity Borsig, Lubor Abstract: Selectins are vascular adhesion molecules that mediate physiological responses such as inflam- mation, immunity and hemostasis. During cancer progression, selectins promote various steps enabling the interactions between tumor cells and the blood constituents, including platelets, endothelial cells and leukocytes. Selectins are carbohydrate-binding molecules that bind to sialylated, fucosylated glycan structures. The increased selectin ligand expression on tumor cells correlates with enhanced metastasis and poor prognosis for cancer patients. While, many studies focused on the role of selectin as a mediator of tumor cell adhesion and extravasation during metastasis, there is evidence for selectins to activate signaling cascade that regulates immune responses within a tumor microenvironment. L-Selectin binding induces activation of leukocytes, which can be further modulated by selectin-mediated interactions with platelets and endothelial cells. Selectin ligand on leukocytes, PSGL-1, triggers intracellular signaling in leukocytes that are induced through platelet’s P-selectin or endothelial E-selectin binding. In this review, I summarize the evidence for selectin-induced immune modulation in cancer progression that represents a possible target for controlling tumor immunity. DOI: https://doi.org/10.1093/glycob/cwx105 Posted at the Zurich Open Repository and -

Molecular and Clinical Characterization of LAG3 in Breast Cancer Through 2994 Samples
Molecular and Clinical Characterization of LAG3 in Breast Cancer Through 2994 Samples Qiang Liu Chinese Academy of Medical Sciences & Peking Union Medical College Yihang Qi ( [email protected] ) Chinese Academy of Medical Sciences and Peking Union Medical College https://orcid.org/0000-0001- 7589-0333 Jie Zhai Chinese Academy of Medical Sciences & Peking Union Medical College Xiangyi Kong Chinese Academy of Medical Sciences & Peking Union Medical College Xiangyu Wang Chinese Academy of Medical Sciences & Peking Union Medical College Yi Fang Chinese Academy of Medical Sciences & Peking Union Medical College Jing Wang Chinese Academy of Medical Sciences & Peking Union Medical College Research Keywords: Cancer immunotherapy, CD223, LAG3, Immune response, Inammatory activity Posted Date: June 19th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-36422/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/33 Abstract Background Despite the promising impact of cancer immunotherapy targeting CTLA4 and PD1/PDL1, a large number of cancer patients fail to respond. LAG3 (Lymphocyte Activating 3), also named CD233, is a protein Coding gene served as alternative inhibitory receptors to be targeted in the clinic. The impact of LAG3 on immune cell populations and co-regulation of immune response in breast cancer remained largely unknown. Methods To characterize the role of LAG3 in breast cancer, we investigated transcriptome data and associated clinical information derived from a total of 2994 breast cancer patients. Results We observed that LAG3 was closely correlated with major molecular and clinical characteristics, and was more likely to be enriched in higher malignant subtype, suggesting LAG3 was a potential biomarker of triple-negative breast cancer. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

5.01.591 Immune Checkpoint Inhibitors
MEDICAL POLICY – 5.01.591 Immune Checkpoint Inhibitors Effective Date: Sept. 1, 2021 RELATED POLICIES/GUIDELINES: Last Revised: Aug. 10, 2021 5.01.543 General Medical Necessity Criteria for Companion Diagnostics Related Replaces: N/A to Drug Approval 5.01.589 BRAF and MEK Inhibitors Select a hyperlink below to be directed to that section. POLICY CRITERIA | DOCUMENTATION REQUIREMENTS | CODING RELATED INFORMATION | EVIDENCE REVIEW | REFERENCES | HISTORY ∞ Clicking this icon returns you to the hyperlinks menu above. Introduction Chemotherapy, often called chemo, is cancer treatment that uses drugs. Radiation and surgery treat one area of cancer. But chemo usually travels through the bloodstream to treat the whole body. Treating the whole body is called a systemic treatment. Immunotherapy is a new type of cancer treatment that helps the body’s immune cells fight the cancer more effectively. Cancer cells sometimes “hide” from the body’s cells that are designed to search for cells that don’t belong, like cancer cells or bacteria. Immune checkpoint inhibitors are drugs that block the way that cancer cells do this and so help the immune cells find them so they can be stopped. It is one of the ways we can make the environment less friendly to the cancer and slow its growth. Current immunotherapy drugs are complex molecules that must be given through a vein (intravenous). In the future, some may be given by a shot (injection) the patient could inject without help. This policy gives information about immunotherapy drugs and the criteria for when they may be medically necessary. Note: The Introduction section is for your general knowledge and is not to be taken as policy coverage criteria. -

Phyre 2 Results for A1YIY0
Email [email protected] Description A1YIY0 Tue Jul 30 13:19:15 BST Date 2013 Unique Job 1035bc4b501530df ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:cell adhesion Chain: A: PDB Molecule:down syndrome cell adhesion molecule 1 c3dmkA_ Alignment 100.0 14 (dscam) isoform PDBTitle: crystal structure of down syndrome cell adhesion molecule (dscam)2 isoform 1.30.30, n-terminal eight ig domains PDB header:cell adhesion 2 c2om5A_ Alignment 100.0 20 Chain: A: PDB Molecule:contactin 2; PDBTitle: n-terminal fragment of human tax1 PDB header:cell adhesion 3 c3jxaA_ Alignment 100.0 21 Chain: A: PDB Molecule:contactin 4; PDBTitle: immunoglobulin domains 1-4 of mouse cntn4 PDB header:signaling protein/transferase Chain: A: PDB Molecule:tek tyrosine kinase variant; 4 c4k0vA_ 100.0 12 Alignment PDBTitle: structural basis for angiopoietin-1 mediated signaling initiation PDB header:immune system Chain: A: PDB Molecule:natural cytotoxicity triggering receptor 1; 5 c1p6fA_ 100.0 9 Alignment PDBTitle: structure of the human natural cytotoxicity receptor nkp46 PDB header:immune system/receptor 6 c1ollA_ Alignment 99.9 9 Chain: A: PDB Molecule:nk receptor; PDBTitle: extracellular region of the human receptor nkp46 PDB header:viral protein Chain: A: PDB Molecule:hoc head outer capsid protein; 7 c3shsA_ 99.9 18 Alignment PDBTitle: three n-terminal domains of the bacteriophage rb49 highly immunogenic2 outer capsid protein (hoc) PDB header:immune system Chain: E: PDB Molecule:natural -

SLPI and Soluble BTLA As Immunological Markers in Severe Bacterial Infections
SLPI and soluble BTLA as immunological markers in severe bacterial infections To my family Örebro Studies in Medicine 211 ANNA LANGE SLPI and soluble BTLA as immunological markers in severe bacterial infections © Anna Lange, 2020 Title: SLPI and soluble BTLA as immunological markers in severe bacterial infections Publisher: Örebro University 2020 www.oru.se/publikationer Print: Örebro University, Repro 04/2020 ISSN 1652-4063 ISBN 978-91-7529-335-6 Abstract Anna Lange (2020): SLPI and soluble BTLA as immunological markers in severe bacterial infections. Örebro Studies in Medicine 211. Clinical presentation, and outcome of infections are affected by host-, and etiology- (focus of infection and pathogen) related factors. The im- mune response is controlled by a network of regulating pathways. This thesis focuses on Secretory Leukocyte Protease Inhibitor (SLPI), a protease inhibitor with anti-inflammatory properties, and the previously non-studied soluble isoform of B and T lymphocyte attenuator (sBTLA), a membrane-associated regulatory protein. Plasma concentrations of SLPI and sBTLA were assessed in relation to etiology, severity, mortality, and markers of inflammation and immunosuppression, in i) community- acquired pneumonia (CAP) (SLPI), ii) intensive care unit (ICU) treated severe sepsis and septic shock (sBTLA), and iii) dynamically in BSI (SLPI and sBTLA). Main findings were: higher expression of SLPI in pneumonia, com- pared to other sources, higher initial concentrations in Streptococcus pneumoniae, and Staphylococcus aureus BSI, compared to Escherichia coli BSI, and higher SLPI concentrations in sepsis compared to non-septic BSI. Interestingly, men with pneumonia had higher plasma levels of SLPI, both in CAP and BSI. Likewise, sBTLA was associated with severity, but preferentially at higher organ failure scores. -

CD226 T Cells Expressing the Receptors TIGIT and Divergent Phenotypes of Human Regulatory
The Journal of Immunology Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226 Christopher A. Fuhrman,*,1 Wen-I Yeh,*,1 Howard R. Seay,* Priya Saikumar Lakshmi,* Gaurav Chopra,† Lin Zhang,* Daniel J. Perry,* Stephanie A. McClymont,† Mahesh Yadav,† Maria-Cecilia Lopez,‡ Henry V. Baker,‡ Ying Zhang,x Yizheng Li,{ Maryann Whitley,{ David von Schack,x Mark A. Atkinson,* Jeffrey A. Bluestone,‡ and Todd M. Brusko* Regulatory T cells (Tregs) play a central role in counteracting inflammation and autoimmunity. A more complete understanding of cellular heterogeneity and the potential for lineage plasticity in human Treg subsets may identify markers of disease pathogenesis and facilitate the development of optimized cellular therapeutics. To better elucidate human Treg subsets, we conducted direct transcriptional profiling of CD4+FOXP3+Helios+ thymic-derived Tregs and CD4+FOXP3+Helios2 T cells, followed by comparison with CD4+FOXP32Helios2 T conventional cells. These analyses revealed that the coinhibitory receptor T cell Ig and ITIM domain (TIGIT) was highly expressed on thymic-derived Tregs. TIGIT and the costimulatory factor CD226 bind the common ligand CD155. Thus, we analyzed the cellular distribution and suppressive activity of isolated subsets of CD4+CD25+CD127lo/2 T cells expressing CD226 and/or TIGIT. We observed TIGIT is highly expressed and upregulated on Tregs after activation and in vitro expansion, and is associated with lineage stability and suppressive capacity. Conversely, the CD226+TIGIT2 population was associated with reduced Treg purity and suppressive capacity after expansion, along with a marked increase in IL-10 and effector cytokine production. These studies provide additional markers to delineate functionally distinct Treg subsets that may help direct cellular therapies and provide important phenotypic markers for assessing the role of Tregs in health and disease. -

ANGPTL2/LILRB2 Signaling Promotes the Propagation of Lung Cancer Cells
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 25 ANGPTL2/LILRB2 signaling promotes the propagation of lung cancer cells Xiaoye Liu1,2,3,*, Xiaoting Yu4,*, Jingjing Xie5, Mengna Zhan6, Zhuo Yu3, Li Xie3, Hongxiang Zeng3, Feifei Zhang3, Guoqiang Chen1,3, Xianghua Yi4 and Junke Zheng2,3 1 Institute of Health Sciences, Shanghai Institute for Biological Sciences, University of Chinese Academy of Science, Chinese Academy of Sciences and Shanghai Jiao Tong University School of Medicine, Shanghai, China 2 Hongqiao International Institute of Medicine, Shanghai Tongren Hospital, Faculty of Basic Medicine, Shanghai Jiao Tong University School of Medicine, Shanghai, China 3 Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai, China 4 Department of Pathology, Tongji Hospital, Tongji University School of Medicine, Shanghai, China 5 Bingzhou Medical University, Taishan Scholar Program, Yantai, China 6 Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China * These authors have contributed equally to this paper Correspondence to: Junke Zheng, email: [email protected] Correspondence to: Xianghua Yi, email: [email protected] Correspondence to: Guoqiang Chen, email: [email protected] Keywords: ANGPTL2/LILRB2 signaling, lung cancer, metastasis, CaMK1 Received: December 31, 2014 Accepted: May 10, 2015 Published: May 20, 2015 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Immune inhibitory receptors expressed on various types of immune cells deliver inhibitory signals that maintain the homeostasis of the immune system. -
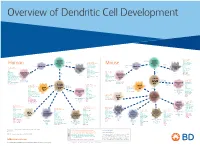
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like,