Defining Molecular Adjuvant Effects on Human B Cell Subsets
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The TNF and TNF Receptor Review Superfamilies: Integrating Mammalian Biology
Cell, Vol. 104, 487±501, February 23, 2001, Copyright 2001 by Cell Press The TNF and TNF Receptor Review Superfamilies: Integrating Mammalian Biology Richard M. Locksley,*²³k Nigel Killeen,²k The receptors and ligands in this superfamily have and Michael J. Lenardo§k unique structural attributes that couple them directly to *Department of Medicine signaling pathways for cell proliferation, survival, and ² Department of Microbiology and Immunology differentiation. Thus, they have assumed prominent ³ Howard Hughes Medical Institute roles in the generation of tissues and transient microen- University of California, San Francisco vironments. Most TNF/TNFR SFPs are expressed in the San Francisco, California 94143 immune system, where their rapid and potent signaling § Laboratory of Immunology capabilities are crucial in coordinating the proliferation National Institute of Allergy and Infectious Diseases and protective functions of pathogen-reactive cells. National Institutes of Health Here, we review the organization of the TNF/TNFR SF Bethesda, Maryland 20892 and how these proteins have been adapted for pro- cesses as seemingly disparate as host defense and or- ganogenesis. In interpreting this large and highly active Introduction area of research, we have focused on common themes that unite the actions of these genes in different tissues. Three decades ago, lymphotoxin (LT) and tumor necro- We also discuss the evolutionary success of this super- sis factor (TNF) were identified as products of lympho- familyÐsuccess that we infer from its expansion across cytes and macrophages that caused the lysis of certain the mammalian genome and from its many indispens- types of cells, especially tumor cells (Granger et al., able roles in mammalian biology. -

Tacrolimus Prevents TWEAK-Induced PLA2R Expression in Cultured Human Podocytes
Journal of Clinical Medicine Article Tacrolimus Prevents TWEAK-Induced PLA2R Expression in Cultured Human Podocytes Leticia Cuarental 1,2, Lara Valiño-Rivas 1,2, Luis Mendonça 3, Moin Saleem 4, Sergio Mezzano 5, Ana Belen Sanz 1,2 , Alberto Ortiz 1,2,* and Maria Dolores Sanchez-Niño 1,2,* 1 IIS-Fundacion Jimenez Diaz, Universidad Autonoma de Madrid, Fundacion Renal Iñigo Alvarez de Toledo-IRSIN, 28040 Madrid, Spain; [email protected] (L.C.); [email protected] (L.V.-R.); [email protected] (A.B.S.) 2 Red de Investigación Renal (REDINREN), Fundacion Jimenez Diaz, 28040 Madrid, Spain 3 Nephrology Department, Centro Hospitalar Universitário São João, 4200-319 Porto, Portugal; [email protected] 4 Bristol Renal, University of Bristol, Bristol BS8 1TH, UK; [email protected] 5 Laboratorio de Nefrologia, Facultad de Medicina, Universidad Austral de Chile, 5090000 Valdivia, Chile; [email protected] * Correspondence: [email protected] (A.O.); [email protected] (M.D.S.-N.); Tel.: +34-91-550-48-00 (A.O. & M.D.S.-N.) Received: 29 May 2020; Accepted: 7 July 2020; Published: 10 July 2020 Abstract: Primary membranous nephropathy is usually caused by antibodies against the podocyte antigen membrane M-type phospholipase A2 receptor (PLA2R). The treatment of membranous nephropathy is not fully satisfactory. The calcineurin inhibitor tacrolimus is used to treat membranous nephropathy, but recurrence upon drug withdrawal is common. TNF superfamily members are key mediators of kidney injury. We have now identified key TNF receptor superfamily members in podocytes and explored the regulation of PLA2R expression and the impact of tacrolimus. -

Cytokine Nomenclature
RayBiotech, Inc. The protein array pioneer company Cytokine Nomenclature Cytokine Name Official Full Name Genbank Related Names Symbol 4-1BB TNFRSF Tumor necrosis factor NP_001552 CD137, ILA, 4-1BB ligand receptor 9 receptor superfamily .2. member 9 6Ckine CCL21 6-Cysteine Chemokine NM_002989 Small-inducible cytokine A21, Beta chemokine exodus-2, Secondary lymphoid-tissue chemokine, SLC, SCYA21 ACE ACE Angiotensin-converting NP_000780 CD143, DCP, DCP1 enzyme .1. NP_690043 .1. ACE-2 ACE2 Angiotensin-converting NP_068576 ACE-related carboxypeptidase, enzyme 2 .1 Angiotensin-converting enzyme homolog ACTH ACTH Adrenocorticotropic NP_000930 POMC, Pro-opiomelanocortin, hormone .1. Corticotropin-lipotropin, NPP, NP_001030 Melanotropin gamma, Gamma- 333.1 MSH, Potential peptide, Corticotropin, Melanotropin alpha, Alpha-MSH, Corticotropin-like intermediary peptide, CLIP, Lipotropin beta, Beta-LPH, Lipotropin gamma, Gamma-LPH, Melanotropin beta, Beta-MSH, Beta-endorphin, Met-enkephalin ACTHR ACTHR Adrenocorticotropic NP_000520 Melanocortin receptor 2, MC2-R hormone receptor .1 Activin A INHBA Activin A NM_002192 Activin beta-A chain, Erythroid differentiation protein, EDF, INHBA Activin B INHBB Activin B NM_002193 Inhibin beta B chain, Activin beta-B chain Activin C INHBC Activin C NM005538 Inhibin, beta C Activin RIA ACVR1 Activin receptor type-1 NM_001105 Activin receptor type I, ACTR-I, Serine/threonine-protein kinase receptor R1, SKR1, Activin receptor-like kinase 2, ALK-2, TGF-B superfamily receptor type I, TSR-I, ACVRLK2 Activin RIB ACVR1B -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -

Human TNFRSF18 ELISA Kit (ARG81453)
Product datasheet [email protected] ARG81453 Package: 96 wells Human TNFRSF18 ELISA Kit Store at: 4°C Component Cat. No. Component Name Package Temp ARG81453-001 Antibody-coated 8 X 12 strips 4°C. Unused strips microplate should be sealed tightly in the air-tight pouch. ARG81453-002 Standard 2 X 10 ng/vial 4°C ARG81453-003 Standard/Sample 30 ml (Ready to use) 4°C diluent ARG81453-004 Antibody conjugate 1 vial (100 µl) 4°C concentrate (100X) ARG81453-005 Antibody diluent 12 ml (Ready to use) 4°C buffer ARG81453-006 HRP-Streptavidin 1 vial (100 µl) 4°C concentrate (100X) ARG81453-007 HRP-Streptavidin 12 ml (Ready to use) 4°C diluent buffer ARG81453-008 25X Wash buffer 20 ml 4°C ARG81453-009 TMB substrate 10 ml (Ready to use) 4°C (Protect from light) ARG81453-010 STOP solution 10 ml (Ready to use) 4°C ARG81453-011 Plate sealer 4 strips Room temperature Summary Product Description ARG81453 Human TNFRSF18 ELISA Kit is an Enzyme Immunoassay kit for the quantification of Human TNFRSF18 in serum, plasma (heparin, EDTA) and cell culture supernatants. Tested Reactivity Hu Tested Application ELISA Specificity There is no detectable cross-reactivity with other relevant proteins. Target Name TNFRSF18 Conjugation HRP Conjugation Note Substrate: TMB and read at 450 nm. Sensitivity 31.25 pg/ml Sample Type Serum, plasma (heparin, EDTA) and cell culture supernatants. Standard Range 62.5 - 4000 pg/ml Sample Volume 100 µl www.arigobio.com 1/3 Precision Intra-Assay CV: 6.3% Inter-Assay CV: 7.0% Alternate Names Tumor necrosis factor receptor superfamily member 18; AITR; CD357; CD antigen CD357; Activation- inducible TNFR family receptor; GITR-D; GITR; Glucocorticoid-induced TNFR-related protein Application Instructions Assay Time ~ 5 hours Properties Form 96 well Storage instruction Store the kit at 2-8°C. -
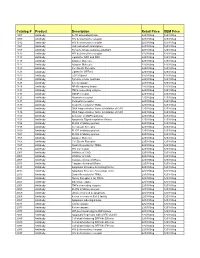
Comprehensive Product List
Catalog # Product Description Retail Price OEM Price 1007 Antibody IL-1R associated kinase 225/100ug 125/100ug 1009 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1012 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1021 Antibody JAK activated transcription 225/100ug 125/100ug 1107 Antibody Tyrosine kinase substrate p62DOK 225/100ug 125/100ug 1112 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1113 Antibody Ligand for DR4 and DR5 225/100ug 125/100ug 1115 Antibody Adapter Molecule 225/100ug 125/100ug 1117 Antibody Adapter Molecule 225/100ug 125/100ug 1120 Antibody Cell Death Receptor 225/100ug 125/100ug 1121 Antibody Ligand for GFRa-2 225/100ug 125/100ug 1123 Antibody CCR3 ligand 225/100ug 125/100ug 1125 Antibody Tyrosine kinase substrate 225/100ug 125/100ug 1128 Antibody A new caspase 225/100ug 125/100ug 1129 Antibody NF-kB inducing kinase 225/100ug 125/100ug 1131 Antibody TNFa converting enzyme 225/100ug 125/100ug 1133 Antibody GDNF receptor 225/100ug 125/100ug 1135 Antibody Neurturin receptor 225/100ug 125/100ug 1137 Antibody Persephin receptor 225/100ug 125/100ug 1139 Antibody Death Receptor for TRAIL 225/100ug 125/100ug 1141 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1148 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1150 Antibody Activator of MAPK pathway 225/100ug 125/100ug 1151 Antibody Apoptosis Signal-regulation Kinase 225/100ug 125/100ug 1156 Antibody FLICE inhibitory protein 225/100ug 125/100ug 1158 Antibody Cell Death Receptor 225/100ug 125/100ug 1159 -

TNFRSF18 (Human) ELISA Kit
TNFRSF18 (Human) ELISA Kit Catalog Number KA1720 96 assays Version: 01 Intended for research use only www.abnova.com I. INTRODUCTION GITR (glucocorticoid induced tumor necrosis factor receptor family related gene) is a type-1 transmembrane protein of 228 amino acids belonging to the TNF and NGF receptor family of proteins. GITR is expressed in normal T-lymphocytes from thymus, spleen. Constitutive expression of a transfected GITR gene induces resistance to apoptosis induced by anti CD3 monoclonal antibodies. The human homolog of GITR is expressed in lymph node and peripheral blood leukocytes. Its expression is up-regulated in human peripheral mononuclear cells mainly after stimulation with antibodies against CD3 and CD28. TNFRSF18 (Human) ELISA Kit is an in vitro enzyme-linked immunosorbent assay for the quantitative measurement of human GITR in cell lysate and tissue lysate. This assay employs an antibody specific for human GITR coated on a 96-well plate. Standards and samples are pipetted into the wells and GITR present in a sample is bound to the wells by the immobilized antibody. The wells are washed and biotinylated anti-human GITR antibody is added. After washing away unbound biotinylated antibody, HRP-conjugated streptavidin is pipetted to the wells. The wells are again washed, a TMB substrate solution is added to the wells and color develops in proportion to the amount of GITR bound. The Stop Solution changes the color from blue to yellow, and the intensity of the color is measured at 450 nm. II. REAGENTS 1. GITR Microplate (Item A): 96 wells (12 strips x 8 wells) coated with anti-human GITR. -

Single-Cell Analysis of Crohn's Disease Lesions Identifies
bioRxiv preprint doi: https://doi.org/10.1101/503102; this version posted December 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy JC Martin1,2,3, G Boschetti1,2,3, C Chang1,2,3, R Ungaro4, M Giri5, LS Chuang5, S Nayar5, A Greenstein6, M. Dubinsky7, L Walker1,2,5,8, A Leader1,2,3, JS Fine9, CE Whitehurst9, L Mbow9, S Kugathasan10, L.A. Denson11, J.Hyams12, JR Friedman13, P Desai13, HM Ko14, I Laface1,2,8, Guray Akturk1,2,8, EE Schadt15,16, S Gnjatic1,2,8, A Rahman1,2,5,8, , M Merad1,2,3,8,17,18*, JH Cho5,17,*, E Kenigsberg1,15,16,17* 1 Precision Immunology Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 2 Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 3 Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 4 The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York City, NY 10029, USA. 5 Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 6 Department of Colorectal Surgery, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA 7 Department of Pediatrics, Susan and Leonard Feinstein IBD Clinical Center, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. -
Adipocytes As Immune Cells: Differential Expression of TWEAK
Adipocytes as Immune Cells: Differential Expression of TWEAK, BAFF, and APRIL and Their Receptors (Fn14, BAFF-R, TACI, and BCMA) at Different Stages of Normal This information is current as and Pathological Adipose Tissue of September 26, 2021. Development Vassilia-Ismini Alexaki, George Notas, Vassiliki Pelekanou, Marilena Kampa, Maria Valkanou, Panayiotis Theodoropoulos, Efstathios N. Stathopoulos, Andreas Tsapis Downloaded from and Elias Castanas J Immunol published online 14 October 2009 http://www.jimmunol.org/content/early/2009/10/14/jimmuno l.0901186 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2009/10/13/jimmunol.090118 Material 6.DC1 Why The JI? Submit online. by guest on September 26, 2021 • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2009 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published October 14, 2009, doi:10.4049/jimmunol.0901186 The Journal of Immunology Adipocytes as Immune Cells: Differential Expression of TWEAK, BAFF, and APRIL and Their Receptors (Fn14, BAFF-R, TACI, and BCMA) at Different Stages of Normal and Pathological Adipose Tissue Development1 Vassilia-Ismini Alexaki,* George Notas,* Vassiliki Pelekanou,2* Marilena Kampa,* Maria Valkanou,* Panayiotis Theodoropoulos,† Efstathios N. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Biolegend.Com
Mechanisms of Cell Death TRAIL (TNFSF10) TNF-α Death Receptor 4 (TNFRSF10A/TRAIL-R1) Death Receptor 5 Zombie Dyes (TNFRSF10B/TRAIL-R2) Propidium Iodide (PI) BAT1, TIM-4 TNF RI (TNFRSF1A) 7-Amino-Actinomycin (7-AAD) MER TNF RII (TNFRSF1B) FAS-L GAS6 (TNFSF6/CD178) TRAIL (TNFSF10) Apoptotic Cell Death Domain Zombie Dyes Phosphatidylserine K63 Ubiquitin NH2 Removal ICAM3? ROCK1 NH CD14 2 Eat-Me Signals FAS Death Inducing Cytoskeletal Rearrangement, (TNFRSF6/CD95) Signaling Complex (DISC) TRADD Cytoskeletal Rearrangement, TRADD Decoy Receptor 2 FADD (TNFRSF10D/TRAIL-R4) Actomysin Contraction Engulfment RIP1 TWEAK RIP1 oxLDL (TNFSF12) FADD CIAP1/2 K63 Ubiquitination Blebbing CD36 Death Receptor 3 TWEAK (TNFSF12) PI FADD (TNFRSF25, APO-3) 7-AAD TRAF1 FADD Procaspase 8,10 TRAF 3 Phagocyte FLIP PANX1 Macrophage Monocyte Neutrophil Dendritic Cell Fibroblast Mast Cell Procaspase 8,10 NF-kB TWEAK-R (TNFRSF12A/Fn14) Find-Me Signals Lysophosphocholine C Caspase 8,10 TRAF5 TRAF2 Sphingosine-1-Phosphate G2A? Nucleotides A Decoy TRAIL Receptor R1 (TNFRSF23) Bid Cell Survival ATP, UTP Decoy TRAIL Receptor R2 (TNFRSF22) Sphingosine-1 TRADD Phosphate Receptor Decoy Receptor 1 (TNFRSF10C/TRAIL-R3) Procaspase 3 Proliferation RIP1 G P2y2 t-Bid Bcl-2 T Chemotaxis, Caspase 3 Bcl-2-xL, MCL-1 ? ICAD RIP1 Engulfment Degradation Bax, Bak Oligomerization TRADD Death Receptor 6 Extracellular ATP Bacterial pore-forming toxins TRAIL (TNFSF10) ICAD (TNFRSF21) Monosodium urate crystals Cholesterol crystals Death Receptor DNA Fragmentation Cholera toxin B, Mitochondria