A Comparison of the Cytoplasmic Domains of the Fas Receptor and the P75 Neurotrophin Receptor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Signaling TNF-Like Ligand 1A/Death Receptor 3 the Adaptor Protein
The Adaptor Protein TRADD Is Essential for TNF-Like Ligand 1A/Death Receptor 3 Signaling This information is current as Yelena L. Pobezinskaya, Swati Choksi, Michael J. Morgan, of September 27, 2021. Xiumei Cao and Zheng-gang Liu J Immunol 2011; 186:5212-5216; Prepublished online 18 March 2011; doi: 10.4049/jimmunol.1002374 http://www.jimmunol.org/content/186/9/5212 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2011/03/18/jimmunol.100237 Material 4.DC1 http://www.jimmunol.org/ References This article cites 29 articles, 13 of which you can access for free at: http://www.jimmunol.org/content/186/9/5212.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 27, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology The Adaptor Protein TRADD Is Essential for TNF-Like Ligand 1A/Death Receptor 3 Signaling Yelena L. Pobezinskaya, Swati Choksi, Michael J. Morgan, Xiumei Cao, and Zheng-gang Liu TNFR-associated death domain protein (TRADD) is a key effector protein of TNFR1 signaling. -

The TNF and TNF Receptor Review Superfamilies: Integrating Mammalian Biology
Cell, Vol. 104, 487±501, February 23, 2001, Copyright 2001 by Cell Press The TNF and TNF Receptor Review Superfamilies: Integrating Mammalian Biology Richard M. Locksley,*²³k Nigel Killeen,²k The receptors and ligands in this superfamily have and Michael J. Lenardo§k unique structural attributes that couple them directly to *Department of Medicine signaling pathways for cell proliferation, survival, and ² Department of Microbiology and Immunology differentiation. Thus, they have assumed prominent ³ Howard Hughes Medical Institute roles in the generation of tissues and transient microen- University of California, San Francisco vironments. Most TNF/TNFR SFPs are expressed in the San Francisco, California 94143 immune system, where their rapid and potent signaling § Laboratory of Immunology capabilities are crucial in coordinating the proliferation National Institute of Allergy and Infectious Diseases and protective functions of pathogen-reactive cells. National Institutes of Health Here, we review the organization of the TNF/TNFR SF Bethesda, Maryland 20892 and how these proteins have been adapted for pro- cesses as seemingly disparate as host defense and or- ganogenesis. In interpreting this large and highly active Introduction area of research, we have focused on common themes that unite the actions of these genes in different tissues. Three decades ago, lymphotoxin (LT) and tumor necro- We also discuss the evolutionary success of this super- sis factor (TNF) were identified as products of lympho- familyÐsuccess that we infer from its expansion across cytes and macrophages that caused the lysis of certain the mammalian genome and from its many indispens- types of cells, especially tumor cells (Granger et al., able roles in mammalian biology. -

Cytokine Nomenclature
RayBiotech, Inc. The protein array pioneer company Cytokine Nomenclature Cytokine Name Official Full Name Genbank Related Names Symbol 4-1BB TNFRSF Tumor necrosis factor NP_001552 CD137, ILA, 4-1BB ligand receptor 9 receptor superfamily .2. member 9 6Ckine CCL21 6-Cysteine Chemokine NM_002989 Small-inducible cytokine A21, Beta chemokine exodus-2, Secondary lymphoid-tissue chemokine, SLC, SCYA21 ACE ACE Angiotensin-converting NP_000780 CD143, DCP, DCP1 enzyme .1. NP_690043 .1. ACE-2 ACE2 Angiotensin-converting NP_068576 ACE-related carboxypeptidase, enzyme 2 .1 Angiotensin-converting enzyme homolog ACTH ACTH Adrenocorticotropic NP_000930 POMC, Pro-opiomelanocortin, hormone .1. Corticotropin-lipotropin, NPP, NP_001030 Melanotropin gamma, Gamma- 333.1 MSH, Potential peptide, Corticotropin, Melanotropin alpha, Alpha-MSH, Corticotropin-like intermediary peptide, CLIP, Lipotropin beta, Beta-LPH, Lipotropin gamma, Gamma-LPH, Melanotropin beta, Beta-MSH, Beta-endorphin, Met-enkephalin ACTHR ACTHR Adrenocorticotropic NP_000520 Melanocortin receptor 2, MC2-R hormone receptor .1 Activin A INHBA Activin A NM_002192 Activin beta-A chain, Erythroid differentiation protein, EDF, INHBA Activin B INHBB Activin B NM_002193 Inhibin beta B chain, Activin beta-B chain Activin C INHBC Activin C NM005538 Inhibin, beta C Activin RIA ACVR1 Activin receptor type-1 NM_001105 Activin receptor type I, ACTR-I, Serine/threonine-protein kinase receptor R1, SKR1, Activin receptor-like kinase 2, ALK-2, TGF-B superfamily receptor type I, TSR-I, ACVRLK2 Activin RIB ACVR1B -

Structural and Functional Diversity of Caspase Homologues in Non-Metazoan Organisms
Protoplasma DOI 10.1007/s00709-017-1145-5 REVIEW ARTICLE Structural and functional diversity of caspase homologues in non-metazoan organisms Marina Klemenčič1,2 & Christiane Funk1 Received: 1 June 2017 /Accepted: 5 July 2017 # The Author(s) 2017. This article is an open access publication Abstract Caspases, the proteases involved in initiation and supports the role of metacaspases and orthocaspases as im- execution of metazoan programmed cell death, are only pres- portant contributors to cell homeostasis during normal physi- ent in animals, while their structural homologues can be ological conditions or cell differentiation and ageing. found in all domains of life, spanning from simple prokary- otes (orthocaspases) to yeast and plants (metacaspases). All members of this wide protease family contain the p20 do- Keywords Algae . Cyanobacteria . Cell death . Cysteine main, which harbours the catalytic dyad formed by the two protease . Metacaspase . Orthocaspase amino acid residues, histidine and cysteine. Despite the high structural similarity of the p20 domain, metacaspases and orthocaspases were found to exhibit different substrate speci- ficities than caspases. While the former cleave their substrates Introduction after basic amino acid residues, the latter accommodate sub- strates with negative charge. This observation is crucial for BOut of life’s school of war: What does not destroy me, the re-evaluation of non-metazoan caspase homologues being makes me stronger.^ wrote the German philosopher involved in processes of programmed cell death. In this re- Friedrich Nietzsche in his book Twilight of the Idols or view, we analyse the structural diversity of enzymes contain- how to philosophize with a hammer. Even though ing the p20 domain, with focus on the orthocaspases, and reformatted to more common use, this phrase has been summarise recent advances in research of orthocaspases and used to describe the dual nature of caspase homologues metacaspases of cyanobacteria, algae and higher plants. -

Comparative Study of Apoptosis-Related Gene Loci in Human, Mouse and Rat Genomes
ISSN 1672-9145 Acta Biochimica et Biophysica Sinica 2005, 37(5): 341–348 CN 31-1940/Q Comparative Study of Apoptosis-related Gene Loci in Human, Mouse and Rat Genomes Yan-Bin YIN, Yong ZHANG, Peng YU, Jing-Chu LUO, Ying JIANG*, and Song-Gang LI* Center of Bioinformatics, National Laboratory of Genetic Engineering and Protein Engineering, College of Life Sciences, Peking University, Beijing 100871, China Downloaded from Abstract Many genes are involved in mammalian cell apoptosis pathway. These apoptosis genes often contain characteristic functional domains, and can be classified into at least 15 functional groups, according to previous reports. Using an integrated bioinformatics platform for motif or domain search from three public mammalian proteomes (International Protein Index database for human, mouse, and rat), we system- atically cataloged all of the proteins involved in mammalian apoptosis pathway. By localizing those proteins http://abbs.oxfordjournals.org/ onto the genomes, we obtained a gene locus centric apoptosis gene catalog for human, mouse and rat. Further phylogenetic analysis showed that most of the apoptosis related gene loci are conserved among these three mammals. Interestingly, about one-third of apoptosis gene loci form gene clusters on mammal chromosomes, and exist in the three species, which indicated that mammalian apoptosis gene orders are also conserved. In addition, some tandem duplicated gene loci were revealed by comparing gene loci clusters in the three species. All data produced in this work were stored in a relational database and may be viewed at http://pcas.cbi.pku.edu.cn/database/apd.php. at Institute of Zoology, CAS on November 2, 2011 Key words apoptosis; gene cluster; comparative genomics; bioinformatics Apoptosis is a regulated program by which cells destroy between mouse and human, they revealed that most themselves [1]. -

FLIP and the Death Effector Domain Family
Oncogene (2008) 27, 6216–6227 & 2008 Macmillan Publishers Limited All rights reserved 0950-9232/08 $32.00 www.nature.com/onc REVIEW FLIP and the death effector domain family JW Yu and Y Shi Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ, USA Death effector domains (DEDs) are protein interaction and stress that impinge on the mitochondria or modules found in a number of proteins known to regulate ‘extrinsically’ from extracellular ligands that activate apoptosis from death receptors. The core DED family death receptors at the cell surface. In either scenario, members that orchestrate programmed cell death from each pathway critically relies on the action of a family of death receptors include the adaptor protein FADD, the cysteine proteases known as caspases to initiate and initiator caspases procaspases-8 and -10 and the regulatory execute the cell death process through a two-step protein c-FLIP. Through homotypic DED interactions, cascade (Riedl and Shi, 2004). In the apoptotic these proteins assemble into the death-inducingsignaling proteolytic cascade, the initiator caspases (caspases-2, complex (DISC) to regulate initiator caspase activation -8, -9 and -10) cleave and consequently activate the and launch the apoptotic proteolytic cascade. A consider- effector caspases (caspases-3, -6 and -7), and the effector able body of evidence, however, is revealingthat the same caspases in turn cleave a large array of substrates to core group of DED-containing proteins also paradoxically dismantle and package the cell into apoptotic bodies. promotes survival and proliferation in lymphocytes and Activation of initiator caspases for both the intrinsic possibly other cell types. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
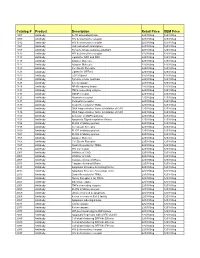
Comprehensive Product List
Catalog # Product Description Retail Price OEM Price 1007 Antibody IL-1R associated kinase 225/100ug 125/100ug 1009 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1012 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1021 Antibody JAK activated transcription 225/100ug 125/100ug 1107 Antibody Tyrosine kinase substrate p62DOK 225/100ug 125/100ug 1112 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1113 Antibody Ligand for DR4 and DR5 225/100ug 125/100ug 1115 Antibody Adapter Molecule 225/100ug 125/100ug 1117 Antibody Adapter Molecule 225/100ug 125/100ug 1120 Antibody Cell Death Receptor 225/100ug 125/100ug 1121 Antibody Ligand for GFRa-2 225/100ug 125/100ug 1123 Antibody CCR3 ligand 225/100ug 125/100ug 1125 Antibody Tyrosine kinase substrate 225/100ug 125/100ug 1128 Antibody A new caspase 225/100ug 125/100ug 1129 Antibody NF-kB inducing kinase 225/100ug 125/100ug 1131 Antibody TNFa converting enzyme 225/100ug 125/100ug 1133 Antibody GDNF receptor 225/100ug 125/100ug 1135 Antibody Neurturin receptor 225/100ug 125/100ug 1137 Antibody Persephin receptor 225/100ug 125/100ug 1139 Antibody Death Receptor for TRAIL 225/100ug 125/100ug 1141 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1148 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1150 Antibody Activator of MAPK pathway 225/100ug 125/100ug 1151 Antibody Apoptosis Signal-regulation Kinase 225/100ug 125/100ug 1156 Antibody FLICE inhibitory protein 225/100ug 125/100ug 1158 Antibody Cell Death Receptor 225/100ug 125/100ug 1159 -

Role of ADAM10 and ADAM17 in Regulating CD137 Function
International Journal of Molecular Sciences Article Role of ADAM10 and ADAM17 in Regulating CD137 Function Jana Seidel 1, Sinje Leitzke 1, Björn Ahrens 1, Maria Sperrhacke 1, Sucharit Bhakdi 2 and Karina Reiss 1,* 1 Department of Dermatology, University of Kiel, 24105 Kiel, Germany; [email protected] (J.S.); [email protected] (S.L.); [email protected] (B.A.); [email protected] (M.S.) 2 Independent Researcher, 24105 Kiel, Germany; [email protected] * Correspondence: [email protected] Abstract: Human CD137 (4-1BB), a member of the TNF receptor family, and its ligand CD137L (4-1BBL), are expressed on immune cells and tumor cells. CD137/CD137L interaction mediates bidirectional cellular responses of potential relevance in inflammatory diseases, autoimmunity and oncology. A soluble form of CD137 exists, elevated levels of which have been reported in patients with rheumatoid arthritis and various malignancies. Soluble CD137 (sCD137) is considered to represent a splice variant of CD137. In this report, however, evidence is presented that A Disintegrin and Metalloproteinase (ADAM)10 and potentially also ADAM17 are centrally involved in its generation. Release of sCD137 by transfected cell lines and primary T cells was uniformly inhibitable by ADAM10 inhibition. The shedding function of ADAM10 can be blocked through inhibition of its interaction with surface exposed phosphatidylserine (PS), and this effectively inhibited sCD137 generation. The phospholipid scramblase Anoctamin-6 (ANO6) traffics PS to the outer membrane and thus modifies ADAM10 function. Overexpression of ANO6 increased stimulated shedding, and hyperactive ANO6 led to maximal constitutive shedding of CD137. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

1471-2164-8-141.Pdf
BMC Genomics BioMed Central Research article Open Access The evolutionary conservation of the core components necessary for the extrinsic apoptotic signaling pathway, in Medaka fish Kazuhiro Sakamaki*1, Masami Nozaki2, Katsuya Kominami1,4 and Yutaka Satou3 Address: 1Department of Animal Development and Physiology, Graduate School of Biostudies, Kyoto University, Kyoto 606-8501, Japan, 2Department of Cell Biology, Research Institute for Microbial Diseases, Osaka University, Suita 565-0871, Japan, 3Department of Zoology, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan and 4Present address: Nihon Schering Research Center, Kobe 650-0047, Japan Email: Kazuhiro Sakamaki* - [email protected]; Masami Nozaki - [email protected]; Katsuya Kominami - [email protected]; Yutaka Satou - [email protected] * Corresponding author Published: 1 June 2007 Received: 16 January 2007 Accepted: 1 June 2007 BMC Genomics 2007, 8:141 doi:10.1186/1471-2164-8-141 This article is available from: http://www.biomedcentral.com/1471-2164/8/141 © 2007 Sakamaki et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Death receptors on the cell surface and the interacting cytosolic molecules, adaptors and initiator caspases, are essential as core components of the extrinsic apoptotic signaling pathway. While the apoptotic machinery governing the extrinsic signaling pathway is well characterized in mammals, it is not fully understood in fish. -

TNFR2 Signaling Regulates the Immunomodulatory Function of Oligodendrocyte Precursor Cells
cells Article TNFR2 Signaling Regulates the Immunomodulatory Function of Oligodendrocyte Precursor Cells Haritha L. Desu 1, Placido Illiano 1, James S. Choi 1, Maureen C. Ascona 1, Han Gao 1,2, Jae K. Lee 1 and Roberta Brambilla 1,3,4,* 1 The Miami Project to Cure Paralysis, Department of Neurological Surgery, University of Miami Miller School of Medicine, Miami, FL 33136, USA; [email protected] (H.L.D.); [email protected] (P.I.); [email protected] (J.S.C.); [email protected] (M.C.A.); [email protected] (H.G.); [email protected] (J.K.L.) 2 Department of Spine Surgery, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou 510630, China 3 Department of Neurobiology Research, Institute of Molecular Medicine, and BRIDGE—Brain Research Inter Disciplinary Guided Excellence, 5000 Odense, Denmark 4 Department of Clinical Research, University of Southern Denmark, 5000 Odense, Denmark * Correspondence: [email protected]; Tel.: +305-243-3567 Abstract: Multiple sclerosis (MS) is a neuroimmune disorder characterized by inflammation, CNS demyelination, and progressive neurodegeneration. Chronic MS patients exhibit impaired remyeli- nation capacity, partly due to the changes that oligodendrocyte precursor cells (OPCs) undergo in response to the MS lesion environment. The cytokine tumor necrosis factor (TNF) is present in the MS-affected CNS and has been implicated in disease pathophysiology. Of the two active forms of TNF, transmembrane (tmTNF) and soluble (solTNF), tmTNF signals via TNFR2 mediating protective and reparative effects, including remyelination, whereas solTNF signals predominantly via TNFR1 Citation: Desu, H.L.; Illiano, P.; Choi, promoting neurotoxicity.