Coevolution Between Nuclear-Encoded DNA Replication, Recombination, and Repair Genes and Plastid Genome Complexity
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

2019 Rare Plants Report
Western Riverside County Multiple Species Habitat Conservation Plan Biological Monitoring Program 2019 Rare Plant Survey Report Brand’s Phacelia (Phacelia stellaris) Little mousetail (Myosurus minimus ) 21 April 2020 i 2019 Rare Plant Survey Report TABLE OF CONTENTS Introduction ......................................................................................................................... 1 Goals and Objectives .......................................................................................................... 1 Methods .............................................................................................................................. 2 Protocol Development ........................................................................................................ 2 Survey Methods .................................................................................................................. 2 Training ............................................................................................................................... 3 Data Analysis ...................................................................................................................... 4 Results ................................................................................................................................. 5 Targeted Surveys ................................................................................................................ 5 Species with Additional Requirements .............................................................................. -

3.4 Biological Resources for the Purpose of This EIR, Biological Resources Comprise Vegetation, Wildlife, Natural Communities, and Wetlands and Other Waters
Impact Analysis Alameda County Community Development Agency Biological Resources 3.4 Biological Resources For the purpose of this EIR, biological resources comprise vegetation, wildlife, natural communities, and wetlands and other waters. Potential biological resource impacts associated with the program and the two individual projects are analyzed. Potential impacts are described quantitatively and qualitatively in Section 3.4.2, Environmental Impacts. This section also identifies specific and detailed measures to avoid, minimize, or compensate for potentially significant impacts on biological resources, where necessary. 3.4.1 Existing Conditions Regulatory Setting Federal Endangered Species Act Pursuant to the federal Endangered Species Act (ESA), USFWS and the National Marine Fisheries Service (NMFS) have authority over projects that may result in take of a species listed as threatened or endangered under the act. Take is defined under the ESA, in part, as killing, harming, or harassing. Under federal regulations, take is further defined to include habitat modification or degradation that results, or is reasonably expected to result, in death or injury to wildlife by significantly impairing essential behavioral patterns, including breeding, feeding, or sheltering. If a likelihood exists that a project would result in take of a federally listed species, either an incidental take permit, under Section 10(a) of the ESA, or a federal interagency consultation, under Section 7 of the ESA, is required. Several federally listed species—vernal pool fairy shrimp (Branchinecta lynchi), longhorn fairy shrimp (Branchinecta longiantenna), vernal pool tadpole shrimp (Lepidurus packardi), California tiger salamander (Ambystoma californiense), California red‐legged frog (Rana draytonii), Alameda whipsnake (Masticophis lateralis euryxanthus), and San Joaquin kit fox (Vulpes macrotis mutica)—have the potential to be affected by activities associated with the Golden Hills and Patterson Pass projects as well as subsequent repowering projects. -

D.1A Floral and Faunal Compendia
Appendix D.1a Floral and Faunal Compendia APPENDIX D.1a FLORAL AND FAUNAL COMPENDIA INTRODUCTION TO FLORAL AND FAUNAL SURVEY Expected site use by wildlife is derived from survey information combined with documented habitat preferences of regional wildlife species, which, whether or not recorded during the survey, are considered likely to include the project area within their range. Habitat designations used in this report are according to the classification system of Holland (1986). Floral taxonomy used in this report follows the Jepson Manual (Hickman 1993), with updates in accordance to the online Jepson Interchange where known. Common plant names, where not available from Munz (1974), are taken from Abrams (1923), Robbins, et al. (1951), Collins (1972), Niehaus and Ripper (1976) and Muns (1983). Vertebrates identified in the field by sight, calls, tracks, scat or other signs are cited according to the nomenclature of Jennings (1983) for amphibians and reptiles; AOU (1983) for birds; and Jones, et al. (1982) for mammals. Butterflies observed or collected in the field were identified with the help of Garth and Tilden (1986) and Tilden and Smith (1986). FLORAL COMPENDIUM1 LEGEND * Nonnative @ Ornamental/Landscape VASCULAR PLANTS CONIFERAE PINACEAE - PINE FAMILY * Pinus halepensis Aleppo Pine ANGIOSPERMAE (DICOTYLEDONS) ANACARDIACEAE - SUMAC FAMILY Malosma laurina laurel sumac Rhus ovata sugar bush * Schinus molle Peruvian pepper-tree Toxicodendron diversilobum poison-oak APOCYNACEAE - DOGBANE FAMILY * Vinca major periwinkle * Nerium oleander oleander ASTERACEAE - SUNFLOWER FAMILY Baccharis pilularis coyote brush Baccharis salicifolia mulefat Helianthus gracilentus slender sunflower * Picris echioides bristly ox-tongue * Silybum marianum milk thistle * Sonchus asper prickly sow-thistle * Sonchus oleraceus common sow-thistle Stephanomeria virgata twiggy wreathplant BRASSICACEAE - MUSTARD FAMILY * Brassica nigra black mustard * Sisymbrium officinale hedge-mustard CAPRIFOLIACEAE - HONEYSUCKLE FAMILY Sambuccus. -

Phylogeny and Historical Biogeography of Geraniaceae In
Systematic Botany (2008), 33(2): pp. 326–342 © Copyright 2008 by the American Society of Plant Taxonomists Phylogeny and Historical Biogeography of Geraniaceae in Relation to Climate Changes and Pollination Ecology Omar Fiz, Pablo Vargas, Marisa Alarcón, Carlos Aedo, José Luis García, and Juan José Aldasoro1 Real Jardín Botanico de Madrid, CSIC, Plaza de Murillo 2, 28014 Madrid, Spain 1Author for correspondence ([email protected]) Communicating Editor: Mark P. Simmons Abstract—Chloroplast (trnL–F and rbcL) sequences were used to reconstruct the phylogeny of Geraniaceae and Hypseocharitaceae. According to these data Hypseocharitaceae and Geraniaceae are monophyletic. Pelargonium and Monsonia are sisters to the largest clade of Geraniaceae, formed by Geranium, Erodium and California. According to molecular dating and dispersal-vicariance analysis, the split of the stem branches of Geraniaceae probably occurred during the Oligocene, in southern Africa or in southern Africa plus the Mediterranean area. However, their diversification occurred during the Miocene, coinciding with the beginning of major aridification events in their distribution areas. An ancestor of the largest clade of Geraniaceae (Geranium, Erodium, and California) colonised a number of habitats in the northern hemisphere and in South American mountain ranges. In summary, the evolution of the Geraniaceae is marked by the dispersal of ancestors from Southern Africa to cold, temperate and often disturbed habitats in the rest of world, where only generalist pollination and facultative autogamy could ensure sufficient seed production and survival. Keywords—autocompatibility, dispersal-vicariance, drought-tolerance, molecular dating, nectaries, P/O indexes. The Geraniaceae are included in the order Geraniales along are characteristic of the Afro-Arabian land mass (Hutchin- with the families Francoaceae, Greyiaceae, Ledocarpaceae, son 1969). -

INVASIVE SPECIES Grass Family (Poaceae) Wild Oats Are Annuals
A PROJECT OF THE SONOMA-MARIN COASTAL PRAIRIE WORKING GROUP INVASIVE SPECIES I NVASIVE A NNUAL P LANTS WILD OATS (AVENA FATUA) AND SLENDER WILD OATS (AVENA BARBATA) - NON-NATIVE Grass Family (Poaceae) Wild oats are annuals. WILD OATS: Are native to Eurasia and North Africa. WILD OAT ECOLOGY Is often dominant or co-dominant in coastal prairie (Ford and Hayes 2007; Sawyer, et al. 2009), Occurs in moist lowland prairies, drier upland prairies and open woodlands (Darris and Gonzalves 2008), Species Interactions: The success of Avena lies in its superior competitive ability: o It has a dense root system. The total root length of a single Avena plant can be from 54.3 miles long (Pavlychenko 1937) to, most likely, twice that long (Dittmer 1937). Wild oats (Avena) in Marin coastal grassland. o It produces allelopathic compounds, Photo by D. (Immel) Jeffery, 2010. chemicals that inhibit the growth of other adjacent plant species. o It has long-lived seeds that can survive for as long as 10 years in the soil (Whitson 2002). Citation: Jeffery (Immel), D., C. Luke, K. Kraft. Last modified February 2020. California’s Coastal Prairie. A project of the Sonoma Marin Coastal Grasslands Working Group, California. Website: www.cnga.org/prairie. Coastal Prairie Described > Species: Invasives: Page 1 of 18 o Pavlychenko (1937) found that, although Avena is a superior competitor when established, it is relatively slow (as compared to cultivated cereal crops wheat, rye and barley) to develop seminal roots in the early growth stages. MORE FUN FACTS ABOUT WILD OATS Avena is Latin for “oat.” The cultivated oat (Avena sativa), also naturalized in California) is thought to be derived from wild oats (Avena fatua) by early humans (Baum and Smith [2011]). -

Parallel Evolution of Highly Conserved Plastid Genome Architecture in Red Seaweeds and Seed Plants
Lee et al. BMC Biology (2016) 14:75 DOI 10.1186/s12915-016-0299-5 RESEARCH ARTICLE Open Access Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants JunMo Lee1, Chung Hyun Cho1, Seung In Park1, Ji Won Choi1, Hyun Suk Song1, John A. West2, Debashish Bhattacharya3† and Hwan Su Yoon1*† Abstract Background: The red algae (Rhodophyta) diverged from the green algae and plants (Viridiplantae) over one billion years ago within the kingdom Archaeplastida. These photosynthetic lineages provide an ideal model to study plastid genome reduction in deep time. To this end, we assembled a large dataset of the plastid genomes that were available, including 48 from the red algae (17 complete and three partial genomes produced for this analysis) to elucidate the evolutionary history of these organelles. Results: We found extreme conservation of plastid genome architecture in the major lineages of the multicellular Florideophyceae red algae. Only three minor structural types were detected in this group, which are explained by recombination events of the duplicated rDNA operons. A similar high level of structural conservation (although with different gene content) was found in seed plants. Three major plastid genome architectures were identified in representatives of 46 orders of angiosperms and three orders of gymnosperms. Conclusions: Our results provide a comprehensive account of plastid gene loss and rearrangement events involving genome architecture within Archaeplastida and lead to one over-arching conclusion: from an ancestral pool of highly rearranged plastid genomes in red and green algae, the aquatic (Florideophyceae) and terrestrial (seed plants) multicellular lineages display high conservation in plastid genome architecture. -

Redalyc.California, a New Genus of Geraniaceae Endemic to The
Anales del Jardín Botánico de Madrid ISSN: 0211-1322 [email protected] Consejo Superior de Investigaciones Científicas España Aldasoro, Juan José; Navarro, Carmen; Vargas, Pablo; Sáez, Llorenç; Aedo, Carlos California, a new genus of Geraniaceae endemic to the southwest of North America Anales del Jardín Botánico de Madrid, vol. 59, núm. 2, 2002, pp. 209-216 Consejo Superior de Investigaciones Científicas Madrid, España Available in: http://www.redalyc.org/articulo.oa?id=55659202 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative CALIFORNIA, A NEW GENUS OF GERANIACEAE ENDEMIC TO THE SOUTHWEST OF NORTH AMERICA by JUAN JOSÉ ALDASORO 1, CARMEN NAVARRO 2, PABLO VARGAS 1, LLORENÇ SÁEZ 3 & CARLOS AEDO 1 1 Real Jardín Botánico, CSIC. Plaza de Murillo, 2. E-28014 Madrid 2 Departamento de Biología Vegetal II, Facultad de Farmacia, Universidad Complutense. E-28040 Madrid 3 Departamento de Botánica, Facultad de Biología, Universidad Autónoma de Bellaterra. E-08193 Barcelona Resumen ALDASORO, J.J., C. NAVARRO, P. VARGAS, LL. SÁEZ & C. AEDO (2002). California, un nuevo género de Geraniaceae endémico del sudoeste de Norteamérica. Anales Jard. Bot. Madrid 59(2): 209-216 (en inglés). Los datos morfológicos permiten distinguir, a nivel de género, Erodium macrophyllum Hook. & Arn. de las especies incluidas en Erodium y Monsonia (Geraniaceae). También los datos obtenidos de la secuencia de ADN cloroplástico (trnL-F) apoyan estas diferencias. Por lo tanto, proponemos un nuevo género monotípico, California Aldas., C. -

Illustration Sources
APPENDIX ONE ILLUSTRATION SOURCES REF. CODE ABR Abrams, L. 1923–1960. Illustrated flora of the Pacific states. Stanford University Press, Stanford, CA. ADD Addisonia. 1916–1964. New York Botanical Garden, New York. Reprinted with permission from Addisonia, vol. 18, plate 579, Copyright © 1933, The New York Botanical Garden. ANDAnderson, E. and Woodson, R.E. 1935. The species of Tradescantia indigenous to the United States. Arnold Arboretum of Harvard University, Cambridge, MA. Reprinted with permission of the Arnold Arboretum of Harvard University. ANN Hollingworth A. 2005. Original illustrations. Published herein by the Botanical Research Institute of Texas, Fort Worth. Artist: Anne Hollingworth. ANO Anonymous. 1821. Medical botany. E. Cox and Sons, London. ARM Annual Rep. Missouri Bot. Gard. 1889–1912. Missouri Botanical Garden, St. Louis. BA1 Bailey, L.H. 1914–1917. The standard cyclopedia of horticulture. The Macmillan Company, New York. BA2 Bailey, L.H. and Bailey, E.Z. 1976. Hortus third: A concise dictionary of plants cultivated in the United States and Canada. Revised and expanded by the staff of the Liberty Hyde Bailey Hortorium. Cornell University. Macmillan Publishing Company, New York. Reprinted with permission from William Crepet and the L.H. Bailey Hortorium. Cornell University. BA3 Bailey, L.H. 1900–1902. Cyclopedia of American horticulture. Macmillan Publishing Company, New York. BB2 Britton, N.L. and Brown, A. 1913. An illustrated flora of the northern United States, Canada and the British posses- sions. Charles Scribner’s Sons, New York. BEA Beal, E.O. and Thieret, J.W. 1986. Aquatic and wetland plants of Kentucky. Kentucky Nature Preserves Commission, Frankfort. Reprinted with permission of Kentucky State Nature Preserves Commission. -

Partial Flora of the Society Islands: Ericaceae to Apocynaceae
SMITHSONIAN CONTRIBUTIONS TO BOTANY NUMBER 17 Partial Flora of the Society Islands: Ericaceae to Apocynaceae Martin Lawrence Grant, F. Raymond Fosberg, and Howard M. Smith SMITHSONIAN INSTITUTION PRESS City of Washington 1974 ABSTRACT Grant, Martin Lawrence, F. Raymond Fosberg, and Howard M. Smith. Partial Flora of the Society Islands: Ericaceae to Apocynaceae. Smithsonian Contri- butions to Botany, number 17, 85 pages, 1974.-Results of a botanical inves- tigation of the Society Islands carried out by Grant in 1930 and 1931, and subsequent work on the material collected and other collections in the U.S. herbaria and other published works are reported herein. This paper is a partial descriptive flora of the Society group with a history of the botanical exploration and investigation of the area. OFFICIALPUBLICATION DATE is handstamped in a limited number of initial copies and is recorded in the Institution’s annual report, Srnithsonian Year. SI PRESS NUMBER 5056. SERIES COVER DESIGN: Leaf clearing from the katsura tree Cercidiphyllurn juponicum Siebold and Zuccarini. Library of Congress Cataloging in Publication Data Grant, Martin Lawrence, 1907-1968. Partial flora of the Society Islands: Ericaceae to Apocynaceae. (Smithsonian contributions to botany, no. 17) Supt. of Docs. no.: SI 1.29:17. 1. Botany-Society Islands. I. Fosberg, Francis Raymond, 1908- , joint author. 11. Smith, Howard Malcolm, 1939- , joint author. 111. Title. IV. Series: Smithsonian Institution. Smith- sonian contributions to botany, no. 17. QK1.2747 no. 17 581’.08s [581.9’96’21] 73-22464 For sale by the Superintendent of Documents, US. Government Printing Office Washington, D.C. 20402 Price $1.75 (paper cover) The senior author, after spending almost a year during 1930 and 1931 in the Society Islands, collecting herbarium material and ecological data, worked inten- sively on a comprehensive flora of this archipelago for the next five years. -
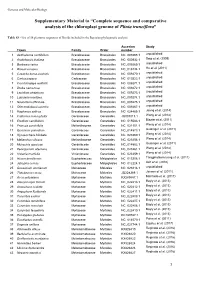
Complete Sequence and Comparative Analysis of the Chloroplast Genome of Plinia Trunciflora”
Genetics and Molecular Biology Supplementary Material to “Complete sequence and comparative analysis of the chloroplast genome of Plinia trunciflora” Table S3 - List of 56 plastome sequences of Rosids included in the Bayesian phylogenetic analysis. Accesion Study Taxon Family Order number 1 Aethionema cordifolium Brassicaceae Brassicales NC_009265.1 unpublished 2 Arabidopsis thaliana Brassicaceae Brassicales NC_000932.1 Sato et al. (1999) 3 Barbarea verna Brassicaceae Brassicales NC_009269.1 unpublished 4 Brassica napus Brassicaceae Brassicales NC_016734.1 Hu et al. (2011) 5 Capsella bursa-pastoris Brassicaceae Brassicales NC_009270.1 unpublished 6 Carica papaya Caricaceae Brassicales NC_010323.1 unpublished 7 Crucihimalaya wallichii Brassicaceae Brassicales NC_009271.1 unpublished 8 Draba nemorosa Brassicaceae Brassicales NC_009272.1 unpublished 9 Lepidium virginicum Brassicaceae Brassicales NC_009273.1 unpublished 10 Lobularia maritima Brassicaceae Brassicales NC_009274.1 unpublished 11 Nasturtium officinale Brassicaceae Brassicales NC_009275.1 unpublished 12 Olimarabidopsis pumila Brassicaceae Brassicales NC_009267.1 unpublished 13 Raphanus sativus Brassicaceae Brassicales NC_024469.1 Jeong et al. (2014) 14 California macrophylla Geraniaceae Geraniales JQ031013.1 Weng et al. (2014) 15 Erodium carvifolium Geraniaceae Geraniales NC_015083.1 Blazier et al. (2011) 16 Francoa sonchifolia Melianthaceae Geraniales NC_021101.1 Weng et al. (2014) 17 Geranium palmatum Geraniaceae Geraniales NC_014573.1 Guisinger et al. (2011) 18 Hypseocharis bilobate Geraniaceae Geraniales NC_023260.1 Weng et al. (2014) 19 Melianthus villosus Melianthaceae Geraniales NC_023256.1 Weng et al. (2014) 20 Monsonia speciose Geraniaceae Geraniales NC_014582.1 Guisinger et al. (2011) 21 Pelargonium alternans Geraniaceae Geraniales NC_023261.1 Weng et al. (2014) 22 Viviania marifolia Vivianiaceae Geraniales NC_023259.1 Weng et al. (2014) 23 Hevea brasiliensis Euphorbiaceae Malpighiales NC_015308.1 Tangphatsornruang et al. -

Downloaded from Genbank on That Full Plastid Genomes Are Not Sufficient to Reject Al- February 28, 2012
Ruhfel et al. BMC Evolutionary Biology 2014, 14:23 http://www.biomedcentral.com/1471-2148/14/23 RESEARCH ARTICLE Open Access From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes Brad R Ruhfel1*, Matthew A Gitzendanner2,3,4, Pamela S Soltis3,4, Douglas E Soltis2,3,4 and J Gordon Burleigh2,4 Abstract Background: Next-generation sequencing has provided a wealth of plastid genome sequence data from an increasingly diverse set of green plants (Viridiplantae). Although these data have helped resolve the phylogeny of numerous clades (e.g., green algae, angiosperms, and gymnosperms), their utility for inferring relationships across all green plants is uncertain. Viridiplantae originated 700-1500 million years ago and may comprise as many as 500,000 species. This clade represents a major source of photosynthetic carbon and contains an immense diversity of life forms, including some of the smallest and largest eukaryotes. Here we explore the limits and challenges of inferring a comprehensive green plant phylogeny from available complete or nearly complete plastid genome sequence data. Results: We assembled protein-coding sequence data for 78 genes from 360 diverse green plant taxa with complete or nearly complete plastid genome sequences available from GenBank. Phylogenetic analyses of the plastid data recovered well-supported backbone relationships and strong support for relationships that were not observed in previous analyses of major subclades within Viridiplantae. However, there also is evidence of systematic error in some analyses. In several instances we obtained strongly supported but conflicting topologies from analyses of nucleotides versus amino acid characters, and the considerable variation in GC content among lineages and within single genomes affected the phylogenetic placement of several taxa. -

Checklist of the Vascular Plants of San Diego County 5Th Edition
cHeckliSt of tHe vaScUlaR PlaNtS of SaN DieGo coUNty 5th edition Pinus torreyana subsp. torreyana Downingia concolor var. brevior Thermopsis californica var. semota Pogogyne abramsii Hulsea californica Cylindropuntia fosbergii Dudleya brevifolia Chorizanthe orcuttiana Astragalus deanei by Jon P. Rebman and Michael G. Simpson San Diego Natural History Museum and San Diego State University examples of checklist taxa: SPecieS SPecieS iNfRaSPecieS iNfRaSPecieS NaMe aUtHoR RaNk & NaMe aUtHoR Eriodictyon trichocalyx A. Heller var. lanatum (Brand) Jepson {SD 135251} [E. t. subsp. l. (Brand) Munz] Hairy yerba Santa SyNoNyM SyMBol foR NoN-NATIVE, NATURaliZeD PlaNt *Erodium cicutarium (L.) Aiton {SD 122398} red-Stem Filaree/StorkSbill HeRBaRiUM SPeciMeN coMMoN DocUMeNTATION NaMe SyMBol foR PlaNt Not liSteD iN THE JEPSON MANUAL †Rhus aromatica Aiton var. simplicifolia (Greene) Conquist {SD 118139} Single-leaF SkunkbruSH SyMBol foR StRict eNDeMic TO SaN DieGo coUNty §§Dudleya brevifolia (Moran) Moran {SD 130030} SHort-leaF dudleya [D. blochmaniae (Eastw.) Moran subsp. brevifolia Moran] 1B.1 S1.1 G2t1 ce SyMBol foR NeaR eNDeMic TO SaN DieGo coUNty §Nolina interrata Gentry {SD 79876} deHeSa nolina 1B.1 S2 G2 ce eNviRoNMeNTAL liStiNG SyMBol foR MiSiDeNtifieD PlaNt, Not occURRiNG iN coUNty (Note: this symbol used in appendix 1 only.) ?Cirsium brevistylum Cronq. indian tHiStle i checklist of the vascular plants of san Diego county 5th edition by Jon p. rebman and Michael g. simpson san Diego natural history Museum and san Diego state university publication of: san Diego natural history Museum san Diego, california ii Copyright © 2014 by Jon P. Rebman and Michael G. Simpson Fifth edition 2014. isBn 0-918969-08-5 Copyright © 2006 by Jon P.