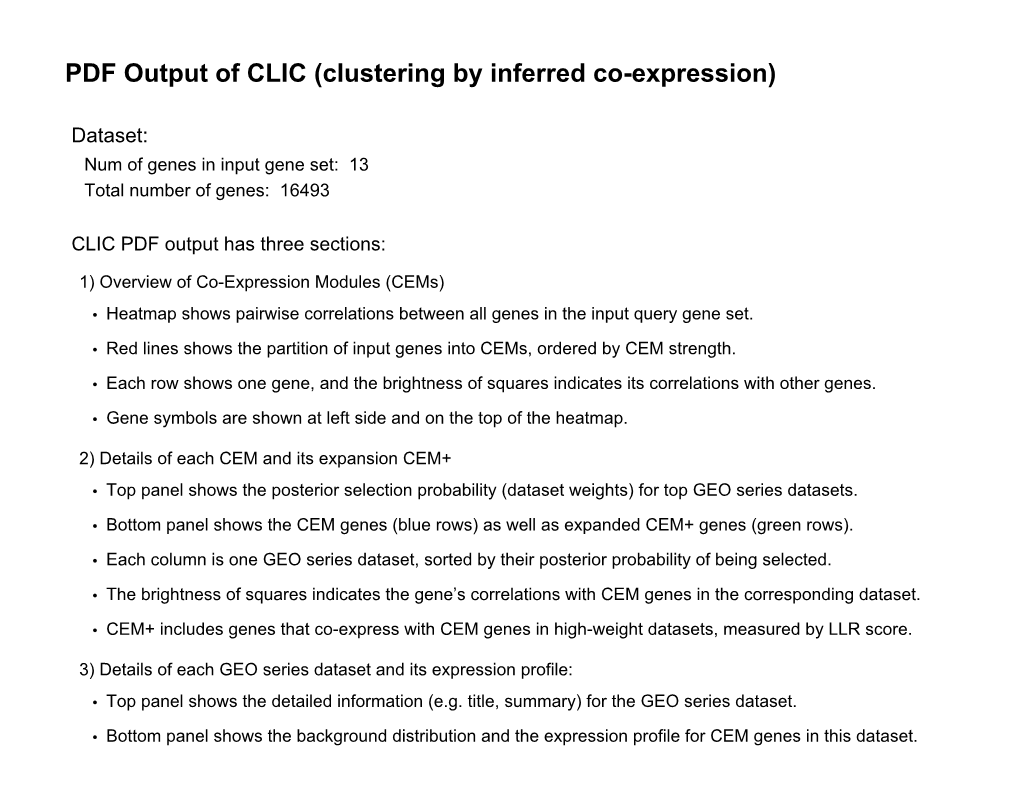
PDF Output of CLIC (Clustering by Inferred Co-Expression)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cellular and Molecular Investigations of Undiagnosed Neurometabolic
CELLULAR AND MOLECULAR INVESTIGATIONS OF UNDIAGNOSED NEUROMETABOLIC DISORDERS Submitted in application for award of Doctor of Philosophy (PhD) Emma Reid Centre for Translational Omics Genetics and Genomic Medicine Institute of Child Health University College London August 2016 DECLARATION I, Emma Reid, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Where experimental or analytical work has been completed by others, this has been stated in the relevant section of this thesis. However, these instances are also listed below: • Alignment of sequencing reads to the reference genome, variant calling (Section 2.4.11) and ExomeDepth analysis (Section 3.3.3) of the gene panel sequencing data was performed by Dr Chris Boustred (NE Thames Regional Genetics Service, GOSH, UK). • Whole exome sequencing of patients X, 1, 4 and 5 was outsourced to BGI Genomics Hong Kong and the resulting raw data files were processed and aligned by Dr Chela James (GOSgene, ICH, UK) (Section 2.5). • Whole exome sequencing of patient Y and processing of the resulting raw data was performed by Dr Olaf Bodamer (University of Miami, USA) (Section 6.4.1). • Homozygosity mapping and whole exome sequencing of index PROSC family was performed by Dr Niklas Darin (The Queen Silvia Children’s Hospital, Sweden) (Section 7.1.4). • Sanger sequencing of additional PROSC and PNPO-deficient patients was performed by Dr Philippa Mills (ICH, UK) (Section 7.2). • Sample processing for electron microscopy analysis was carried out by Elizabeth Latimer- Bowman (Histopathology Department, GOSH, UK) and imaging was performed by Glenn Anderson (Histopathology Department, GOSH, UK) (Section 2.14.2). -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Datasheet: VMA00460 Product Details
Datasheet: VMA00460 Description: MOUSE ANTI PROSC Specificity: PROSC Format: Purified Product Type: PrecisionAb™ Monoclonal Clone: 3A6 Isotype: IgG1 Quantity: 100 µl Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Species Cross Reacts with: Mouse Reactivity N.B. Antibody reactivity and working conditions may vary between species. Product Form Purified IgG - liquid Preparation Purified IgG prepared by affinity chromatography on Protein G from ascites Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers 1% Bovine Serum Albumin 50% Glycerol Immunogen Full length recombinant protein of human PROSC produced in E. Coli. External Database Links UniProt: O94903 Related reagents Entrez Gene: 11212 PROSC Related reagents Page 1 of 2 Specificity Mouse anti Human PROSC antibody recognizes PROSC also known as proline synthase co-transcribed bacterial homolog protein. PROSC is a ubiquitoously expressed 275 amino acid cytoplasmic protein highly conserved between divergent species (Ikegawa 1999). Mouse anti Human PROSC antibody detects a band of 30 kDa. -

PDXK Mutations Cause Polyneuropathy Responsive to Pyridoxal 50-Phosphate Supplementation
RESEARCH ARTICLE PDXK Mutations Cause Polyneuropathy Responsive to Pyridoxal 50-Phosphate Supplementation Viorica Chelban, MD, MSc ,1,2 Matthew P. Wilson, PhD,3* Jodi Warman Chardon, MD,4,5,6* Jana Vandrovcova, PhD,1* M. Natalia Zanetti, PhD,7 Eleni Zamba-Papanicolaou, MD,8,9 Stephanie Efthymiou, MSc,1 Simon Pope, PhD,10 Maria R. Conte, PhD,11 Giancarlo Abis, PhD ,11 Yo-Tsen Liu, PhD ,12,13,14 Eloise Tribollet, MS,1 Nourelhoda A. Haridy, MD,1,15 Juan A. Botía, PhD,16,17 Mina Ryten, PhD,16,18 Paschalis Nicolaou, PhD,8,9 Anna Minaidou, PhD,8,9 Kyproula Christodoulou, PhD,8,9 Kristin D. Kernohan, PhD,6,19 Alison Eaton, MD,6 Matthew Osmond, MSc,6 Yoko Ito, PhD,6 Pierre Bourque, MD,4,5 James E. C. Jepson, PhD,7 Oscar Bello, PhD,7 Fion Bremner, MD,20 Carla Cordivari, MD,21 Mary M. Reilly, MD, FRCP, FRCPI, PhD,1 Martha Foiani, MSc,21,22 Amanda Heslegrave, PhD,22,23 Henrik Zetterberg, PhD,22,23,24,25 Simon J. R. Heales, PhD,10 Nicholas W. Wood, PhD,1,26 James E. Rothman, PhD,7,27 Kym M. Boycott, MD, PhD,6 † † † Philippa B. Mills, PhD,3 Peter T. Clayton, PhD,3 and Henry Houlden, PhD,1,26 for the Care4Rare Canada Consortium and the SYNaPS Study Group6 View this article online at wileyonlinelibrary.com. DOI: 10.1002/ana.25524 Received Oct 1, 2018, and in revised form Jun 5, 2019. Accepted for publication Jun 7, 2019. Address correspondence to Dr Chelban and Prof Houlden, Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, United Kingdom. -

Implications for the 8P12-11 Genomic Locus in Breast Cancer Diagnostics and Therapy: Eukaryotic Initiation Factor 4E-Binding Protein, EIF4EBP1, As an Oncogene
Medical University of South Carolina MEDICA MUSC Theses and Dissertations 2018 Implications for the 8p12-11 Genomic Locus in Breast Cancer Diagnostics and Therapy: Eukaryotic Initiation Factor 4E-Binding Protein, EIF4EBP1, as an Oncogene Alexandria Colett Rutkovsky Medical University of South Carolina Follow this and additional works at: https://medica-musc.researchcommons.org/theses Recommended Citation Rutkovsky, Alexandria Colett, "Implications for the 8p12-11 Genomic Locus in Breast Cancer Diagnostics and Therapy: Eukaryotic Initiation Factor 4E-Binding Protein, EIF4EBP1, as an Oncogene" (2018). MUSC Theses and Dissertations. 295. https://medica-musc.researchcommons.org/theses/295 This Dissertation is brought to you for free and open access by MEDICA. It has been accepted for inclusion in MUSC Theses and Dissertations by an authorized administrator of MEDICA. For more information, please contact [email protected]. Implications for the 8p12-11 Genomic Locus in Breast Cancer Diagnostics and Therapy: Eukaryotic Initiation Factor 4E-Binding Protein, EIF4EBP1, as an Oncogene by Alexandria Colett Rutkovsky A dissertation submitted to the faculty of the Medical University of South Carolina in fulfillment of the requirements for the degree of Doctor of Philosophy in the College of Graduate Studies Biomedical Sciences Department of Pathology and Laboratory Medicine 2018 Approved by: Chairman, Advisory Committee Stephen P. Ethier Co-Chairman Robin C. Muise-Helmericks Amanda C. LaRue Victoria J. Findlay Elizabeth S. Yeh Copyright by Alexandria Colett Rutkovsky 2018 All Rights Reserved This work is dedicated to the fight against cancer. ACKNOWLEDGEMENTS I would like to acknowledge the Hollings Cancer Center and many departments at the Medical University of South Carolina for investing the resources and training required to complete the research presented. -

Potential Oncogenes Within the 8P12-P11 Amplicon
Potential oncogenes within the 8p12-p11 amplicon Identification and functional testing in breast cancer cell lines Edda Olgudóttir Thesis for the degree of Master of Science University of Iceland Faculty of Medicine School of Health Science Hugsanleg æxlisgen á 8p12-p11 mögnunarsvæðinu Æxlisgen tilgreind og prófuð í brjóstakrabbameinsfrumulínum Edda Olgudóttir Ritgerð til meistaragráðu í Líf- og læknavísindum Leiðbeinendur: Inga Reynisdóttir og Rósa Björk Barkardóttir Meistaranámsnefnd: Inga Reynisdóttir, Rósa Björk Barkardóttir og Guðmundur Hrafn Guðmundsson Læknadeild Heilbrigðisvísindasvið Háskóla Íslands Febrúar 2012 Potential oncogenes within the 8p12-p11 amplicon Identification and functional testing in breast cancer cell lines Edda Olgudóttir Thesis for the degree of Master of Science Supervisor: Inga Reynisdóttir and Rósa Björk Barkardóttir Masters committee: Inga Reynisdóttir, Rósa Björk Barkardóttir and Guðmundur Hrafn Guðmundsson Faculty of Medicine School of Health Sciences February 2012 Ritgerð þessi er til meistaragráðu í Líf- og læknavísindum og er óheimilt að afrita ritgerðina á nokkurn hátt nema með leyfi rétthafa. © Edda Olgudóttir 2012 Prentun: Háskólaprent Staður, Ísland 2012 Ágrip Orsakir brjóstakrabbameins eru margþættar og margar þeirra leiða til brenglana í erfðaefninu. Algengar erfðabrenglanir í brjóstakrabbameini eru tap eða magnanir á litningasvæðum. Svæði sem eru gjörn á að tapast innihalda svokölluð æxlisbæligen en svæði sem magnast innihalda æxlisgen, gen sem ýta undir æxlisvöxt. Æxlisgen sem eru staðsett á mögnunarsvæði eru oft yfirtjáð samhliða mögnun þeirra og hafa þar af leiðandi aukna virkni. Vegna þessa má líta svo á að æxlisgenin drífi mögnunina áfram og þau séu því markgen mögnunarinnar. Litningasvæðið 8p12-p11 finnst magnað í u.þ.b. 15-20% brjóstakrabbameina. Hingað til hefur einungis tekist að skilgreina markgen mögnunarinnar í einum undirflokk brjóstakrabbameina, luminal B undirflokk. -

Research 1..8
Article pubs.acs.org/jpr Proteome Atlas of Human Chromosome 8 and Its Multiple 8p Deficiencies in Tumorigenesis of the Stomach, Colon, and Liver † ○ † ○ ‡ § ○ ∥ ○ † † † Yang Zhang, , Guoquan Yan, , Linhui Zhai, , , Shaohang Xu, , Huali Shen, Jun Yao, Feifei Wu, † # † † † ‡ § ‡ § Liqi Xie, Hailin Tang, Hongxiu Yu, Mingqi Liu, Pengyuan Yang, Ping Xu, , Chengpu Zhang, , ‡ § ‡ § ‡ § ‡ § ‡ § ∥ ⊥ ∥ Liwei Li, , Cheng Chang, , Ning Li, , Songfeng Wu, , Yunping Zhu, , Quanhui Wang, , Bo Wen, ∥ ∥ ∥ □ † ∥ ⊥ ‡ § Liang Lin, Yinzhu Wang, Guiyan Zheng, Lanping Zhou, Haojie Lu,*, Siqi Liu,*, , Fuchu He,*, , † and Fan Zhong*, † Institutes of Biomedical Sciences and Department of Chemistry, Fudan University, Shanghai 200032, China ‡ State Key Laboratory of Proteomics, Beijing Proteome Research Center, Beijing Institute of Radiation Medicine, Beijing 102206, China § National Engineering Research Center for Protein Drugs, Beijing 102206, China ∥ BGI-Shenzhen, Shenzhen 518083, China ⊥ Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100029, China # College of Mechanical & Electronic Engineering and Automatization, National University of Defense Technology, Changsha 410073, China □ State Key Laboratory of Molecular Oncology, Cancer Institute & Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100021, China *S Supporting Information ABSTRACT: Chromosome 8, a medium-length euchromatic unit in humans that has an extraordinarily high mutation rate, can be detected not only in evolution but also in multiple -

WO 2015/149034 A2 1 October 2015 (01.10.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/149034 A2 1 October 2015 (01.10.2015) P O P C T (51) International Patent Classification: (71) Applicant: LIFE TECHNOLOGIES CORPORATION C12Q 1/68 (2006.01) [US/US]; c/o IP Legal Department Docketing, 5791 Van Allen Way, Carlsbad, California 92008 (US). (21) International Application Number: PCT/US2015/023 197 (72) Inventors: RHODES, Daniel; Life Technologies Corpora tion, c/o IP Legal Department Docketing, 579 1 Van Allen (22) International Filing Date: Way, Carlsbad, California 92008 (US). SADIS, Seth; Life 27 March 2015 (27.03.2015) Technologies Corporation, c/o IP Legal Department Dock (25) Filing Language: English eting, 5791 Van Allen Way, Carlsbad, California 92008 (US). WYNGAARD, Peter; Life Technologies Corpora (26) Publication Language: English tion, c/o IP Legal Department Docketing, 579 1 Van Allen (30) Priority Data: Way, Carlsbad, California 92008 (US). KHAZANOV, 61/971,455 27 March 2014 (27.03.2014) US Nikolay; Life Technologies Corporation, c/o IP Legal De 61/993,732 15 May 2014 (15.05.2014) US partment Docketing, 5791 Van Allen Way, Carlsbad, Cali 62/004,727 29 May 2014 (29.05.2014) us fornia 92008 (US). BANDLA, Santhoshi; Life Technolo 62/092,898 17 December 2014 (17. 12.2014) us gies Corporation, c/o IP Legal Department Docketing, 5791 Van Allen Way, Carlsbad, California 92008 (US). [Continued on nextpage] (54) Title: GENE FUSIONS AND GENE VARIANTS ASSOCIATED WITH CANCER (57) Abstract: The disclosure provides gene fusions, gene variants, and novel as RNASeq sociations with disease states, as well as kits, probes, and methods of using the (.bam) same. -

Antibodies Reactive to Non-HLA Antigens in Transplant Glomerulopathy
CLINICAL RESEARCH www.jasn.org Antibodies Reactive to Non-HLA Antigens in Transplant Glomerulopathy Rajani Dinavahi,*† Ajish George,‡ Anne Tretin,* Enver Akalin,*† Scott Ames,† Jonathan S. Bromberg,† Graciela DeBoccardo,*† Nicholas DiPaola,§ Susan M. Lerner,† ʈ Anita Mehrotra,* Barbara T. Murphy,*† Tibor Nadasdy, Estela Paz-Artal,¶ Daniel R. Salomon,** Bernd Schro¨ppel,*† Vinita Sehgal,*† Ravi Sachidanandam,‡ and Peter S. Heeger*† *Department of Medicine, †Recanati Miller Transplantation Institute, and ‡Department of Genetics and Genomics ʈ Sciences, Mount Sinai School of Medicine, New York, New York; §Histocompatibility Laboratory and Department of Pathology, Ohio State Medical Center, Columbus, Ohio; ¶Hospital 12 de Octubre, Madrid, Spain; and **The Scripps Research Institute, La Jolla, California ABSTRACT Although T and B cell alloimmunity contribute to transplant injury, autoimmunity directed at kidney- expressed, non-HLA antigens may also participate. Because the specificity, prevalence, and importance of antibodies to non-HLA antigens in late allograft injury are poorly characterized, we used a protein microarray to compare antibody repertoires in pre- and post-transplant sera from several cohorts of patients with and without transplant glomerulopathy. Transplantation routinely induced changes in antibody repertoires, but we did not identify any de novo non-HLA antibodies common to patients with transplant glomerulopathy. The screening studies identified three reactivities present before transplan- tation that persisted after transplant and strongly associated with transplant glomerulopathy. ELISA confirmed that reactivity against peroxisomal-trans-2-enoyl-coA-reductase strongly associated with the development of transplant glomerulopathy in independent validation sets. In addition to providing insight into effects of transplantation on non-HLA antibody repertoires, these results suggest that pretransplant serum antibodies to peroxisomal-trans-2-enoyl-coA-reductase may predict prognosis in kidney transplantation. -

NRG1 Gene Rearrangements in Clinical Breast Cancer: Identification of an Adjacent Novel Amplicon Associated with Poor Prognosis
Oncogene (2005) 24, 7281–7289 & 2005 Nature Publishing Group All rights reserved 0950-9232/05 $30.00 www.nature.com/onc NRG1 gene rearrangements in clinical breast cancer: identification of an adjacent novel amplicon associated with poor prognosis Leah M Prentice1,2, Ashleen Shadeo3, Valia S Lestou1,2,4, Melinda A Miller1,2, Ronald J deLeeuw3, Nikita Makretsov1,2, Dmitry Turbin1,2, Lindsay A Brown1,2, Nicol Macpherson5, Erika Yorida1,2, Maggie CU Cheang1,2, John Bentley1,2, Stephen Chia6, Torsten O Nielsen1,2, C Blake Gilks1,2, Wan Lam3 and David G Huntsman*,1,2 1Department of Pathology and Prostate Centre, Genetic Pathology Evaluation Centre, Vancouver General Hospital, Vancouver, BC, Canada V6H 3Z6; 2British Columbia Cancer Agency, Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, BC, Canada V6H 3Z6; 3Department of Cancer Genetics and Developmental Biology, British Columbia Cancer Research Centre, Vancouver, BC, Canada V5Z 1L3; 4Department of Paediatric Laboratory Medicine, The Hospital for Sick Kids, Toronto, Canada M5G 1X8; 5Department of Medical Oncology, British Columbia Cancer Agency, Victoria, Canada V8R 6V5; 6Department of Medical Oncology, British Columbia Cancer Agency, Vancouver, BC, Canada V5Z 4E6 Rearrangements of the neuregulin (NRG1) gene have been Introduction implicated in breast carcinoma oncogenesis. To determine the frequency and clinical significance of NRG1 aberra- Chromosomal translocations are a common feature of tions in clinical breast tumors,a breast cancer tissue sarcoma, leukemia and lymphoma, but little is known microarray was screened for NRG1 aberrations by about their role in common epithelial malignancies. A fluorescent in situ hybridization (FISH) using a two-color translocation t(12;15) leading to ETV6-NTRK3 gene split-apart probe combination flanking the NRG1 gene. -

Protein Phosphatase and TRAIL Receptor Genes As New Candidate Tumor Genes on Chromosome 8P in Prostate Cancer MAX HORNSTEIN 1, MICHÈLE J
CANCER GENOMICS & PROTEOMICS 5: 123-136 (2008) Protein Phosphatase and TRAIL Receptor Genes as New Candidate Tumor Genes on Chromosome 8p in Prostate Cancer MAX HORNSTEIN 1, MICHÈLE J. HOFFMANN 1, ADRIAN ALEXA 2, MASANORI YAMANAKA 1, MIRKO MÜLLER 1, VOLKER JUNG 3, JÖRG RAHNENFÜHRER 4 and WOLFGANG A. SCHULZ 1* 1Department of Urology, Heinrich Heine University, Düsseldorf; 2Max-Planck Institute for Informatics, 66123 Saarbrücken; 3Department of Urology, Medical University of the Saarland, Homburg; 4Institute for Statistics, University of Dortmund, Dortmund, Germany Abstract. Background: Allelic losses on chromosome 8p location of genes functionally important for their are common in prostate carcinoma, but it is not known development and progression. This conclusion is based on exactly how they contribute to cancer development and the observation that specific chromosomal losses often progression. Materials and Methods: Expression of 12 genes inactivate the second copy of tumor suppressors, whereas located across chromosome 8p, including established tumor chromosomal gains or amplifications lead to the suppressor candidates (CSMD1, DLC1, NKX3.1), and overexpression of oncogenes. For instance, loss of 17p often others from a new microarray-based comparison was signifies the functional inactivation of TP53, whereas gains studied by quantitative RT-PCR in 45 M0 prostate or amplifications at 8q24 are often associated with carcinomas and 13 benign prostate tissues. Results: overexpression of oncogenic MYC. Unfortunately, the Significantly reduced expression was observed for two association between chromosomal changes and the protein phosphatase subunit genes (PPP2CB, PPP3CC) and inactivation or oncogenic activation of individual genes is not two TRAIL decoy receptors (TNFRSF10C/DcR1, always straightforward. Thus, in prostate cancer, it has not TNFRSF10D/DcR2), but not for the three established yet been possible to link the common losses at 8p to the candidates nor for TRAIL death receptor genes. -

Datasheet: VMA00460KT Product Details
Datasheet: VMA00460KT Description: PROSC ANTIBODY WITH CONTROL LYSATE Specificity: PROSC Format: Purified Product Type: PrecisionAb™ Monoclonal Clone: 3A6 Isotype: IgG1 Quantity: 2 Westerns Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Species Cross Reacts with: Mouse Reactivity N.B. Antibody reactivity and working conditions may vary between species. Product Form Purified IgG - liquid Preparation 20μl Mouse monoclonal antibody purified by affinity chromatography on Protein G from ascites Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers 1% Bovine Serum Albumin 50% Glycerol Immunogen Full length recombinant protein of human PROSC produced in E. Coli. External Database Links UniProt: O94903 Related reagents Entrez Gene: 11212 PROSC Related reagents Page 1 of 2 Specificity Mouse anti Human PROSC antibody recognizes PROSC also known as proline synthase co-transcribed bacterial homolog protein. PROSC is a ubiquitoously expressed 275 amino acid cytoplasmic protein highly conserved between divergent species (Ikegawa 1999). Mouse anti Human PROSC antibody detects a band of 30 kDa.