Is a COVID-19 Second Wave Possible in Emilia-Romagna (Italy)? Forecasting a Future Outbreak with Particulate Pollution and Machine Learning
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Camera Di Commercio Della ROMAGNA - FORLI'-CESENA E RIMINI Registro Imprese - Archivio Ufficiale Della CCIAA
Camera di Commercio della ROMAGNA - FORLI'-CESENA e RIMINI Registro Imprese - Archivio ufficiale della CCIAA VISURA DI EVASIONE LIVIA TELLUS ROMAGNA DATI ANAGRAFICI Indirizzo Sede legale FORLI' (FC) PIAZZA AURELIO HOLDING S.P.A. SAFFI 8 CAP 47121 Indirizzo PEC [email protected] Numero REA FO - 323099 Codice fiscale e n.iscr. al 03943760409 Registro Imprese Forma giuridica societa' per azioni La presente visura di evasione è fornita unicamente a riscontro dell'evasione del protocollo dell'istanza. Si ricorda che la visura ufficiale aggiornata dell'impresa è consultabile gratuitamente, da parte del legale rappresentante, tramite il cassetto digitale dell'imprenditore all’indirizzo www.impresa.italia.it Estremi di firma digitale Servizio realizzato da InfoCamere per conto delle Camere di Commercio Italiane Documento n . T 419861469 estratto dal Registro Imprese in data 13/01/2021 Registro Imprese Archivio ufficiale della CCIAA LIVIA TELLUS ROMAGNA HOLDING S.P.A. Documento n . T 419861469 Codice Fiscale 03943760409 estratto dal Registro Imprese in data 13/01/2021 Indice 1 Informazioni da statuto/atto costitutivo .................................... 2 2 Capitale e strumenti finanziari ................................................. 4 3 Soci e titolari di diritti su azioni e quote ................................... 5 4 Amministratori ......................................................................... 12 5 Sindaci, membri organi di controllo ......................................... 14 6 Trasferimenti d'azienda, fusioni, scissioni, -
Rams Remember Recent Rise to Glory
Search for The Westfield News Westfield350.comTheThe Westfield WestfieldNews News Serving Westfield, Southwick, and surrounding Hilltowns “TIME IS THE ONLY WEATHER CRITIC WITHOUT TONIGHT AMBITION.” Partly Cloudy. JOHN STEINBECK Low of 55. www.thewestfieldnews.com VOL. 86 NO. 151 TUESDAY, JUNE 27, 2017 75 cents $1.00 TUESDAY, JUNE 16, 2020 VOL. 89 NO. 143 City Survey on remote auditor learning experience sent to city students gives By AMY PORTER Staff Writer WESTFIELD – The Westfield Public Schools district June 12 sent out a survey to all students about their experience notice with remote learning and thoughts about the fall. By AMY PORTER Superintendent Stefan Staff Writer Czaporowski also wrote a letter WESTFIELD – City Auditor to parents about the importance Christopher Caputo has of the survey and the status of announced he will be leaving back to school planning. A sur- Westfield in the beginning of vey to parents will also be sent July to take a position in a out mid-week. nearby community. In the letter, Czaporowski Caputo, who lives in said that no decisions have Springfield, started in February been made yet on what the of 2019 and will be leaving on return to school will look like July 10. Southwick’s Dan Burnett steals third in Westfield. Caputo said he wasn’t look- base in the 2019 West Division 3 semi- “Westfield, like most school districts, is waiting for a fall ing, but got a call, and couldn’t Southwick’s Josh Lis scores a run against St. final against Taconic at Westfield State guidance memo from Commissioner Riley. -

The Packaging Machinery Cluster in Bologna
Collective Goods in the Local Economy: The Packaging Machinery Cluster in Bologna Paper by Henry Farrell and Ann-Louise Lauridsen March 2001 The debate about the industrial districts of central and north-eastern Italy has evolved over the last 25 years. Initially, many saw them as evidence that small firms could prosper contrary to the arguments of the proponents of big industry. Debate focussed on whether small firm industrial districts had a genuine independent existence, or were the contingent result of large firms’ outsourcing strategies (Brusco 1990, Bagnasco 1977, Bagnasco 1978). This spurred discussion about the role of local and regional government and political parties – small firm success might need services from government, associations, or local networks (Brusco 1982, Trigilia 1986). The difficulties that many industrial districts experienced in the late 1980s and early 1990s, together with the greater flexibility of large firms, led to a second wave of research, which asked whether industrial districts had long term prospects (Harrison 1994, Trigilia 1992, Bellandi 1992, Cooke and Morgan 1994). The most recent literature examines the responses of industrial districts to these challenges; it is clear that many industrial districts have adapted successfully to changing market conditions, but only to the extent that they have changed their modes of internal organisation, and their relationship with the outside world (Amin 1998, Bellandi 1996, Dei Ottati 1996a, Dei Ottati 1996b, Burroni and Trigilia 2001). While these debates have generated important findings, much basic conceptual work remains unfinished. There is still no real consensus about what forces drive evolution in industrial districts and lead to their success or failure. -

{FREE} Emilia-Romagna Travel Guide: Sightseeing, Hotel, Restaurant
EMILIA-ROMAGNA TRAVEL GUIDE: SIGHTSEEING, HOTEL, RESTAURANT & SHOPPING HIGHLIGHTS PDF, EPUB, EBOOK Emily Sutton | 50 pages | 21 Nov 2014 | Createspace | 9781503317666 | English | United States Emilia-Romagna Travel Guide: Sightseeing, Hotel, Restaurant & Shopping Highlights PDF Book Spiaggia Le Palme reviews. Modena is famous for its favorite son Pavarotti. Log in to get trip updates and message other travelers. Learn more - eBay Money Back Guarantee - opens in new window or tab. Posts to:. Rimini centro 1, reviews. Delivery times may vary, especially during peak periods. The Castle Estense sits in the middle of this boulevard. The region is a foodie paradise in a country rewned for its cuisine. Visit store. Charles and St. Travelers to the Emilia Romagna enjoy its flavors and scents through its fine food cultivated from local farms located on its fertile plains. This item will be posted through the Global Shipping Program and includes international tracking. Select a valid country. Be it winter, spring, summer, or autumn there are locally grown seasonal ingredients to be found. The Emilia-Romagna region is a cultural center of Northern Italy. Show more Show less. Explore Province of Rimini. Today the allure of Emilia Romagna sightseeing includes indulging in the food from its fertile plains, enjoying its enchanting scenery, marveling in its art and strolling its history laden cities. Item specifics Condition: Brand new: A new, unread, unused book in perfect condition with no missing or damaged pages. Payment details. Seller posts within 15 days after receiving cleared payment - opens in a new window or tab. Postage and handling. It boasts the longest beach in all Europe. -

The First Diffusion of the Covid-19 Outbreak in Northern Italy
Epidemiol. Methods 2021; 10(s1): 20200047 Mauro Magnoni* The first diffusion of the Covid-19 outbreak in Northern Italy: an analysis based on a simplified version of the SIR model https://doi.org/10.1515/em-2020-0047 Received October 29, 2020; accepted March 10, 2021; published online March 25, 2021 Abstract: In this paper an analysis of the first diffusion of the Covid-19 outbreak occurred in late February 2020 in Northern Italy is presented. In order to study the time evolution of the epidemic it was decided to analyze in particular as the most relevant variable the number of hospitalized people, considered as the less biased proxy of the real number of infected people. An approximate solution of the infected equation was found from a simplified version of the SIR model. This solution was used as a tool for the calculation ofthe basic reproduction number R0 in the early phase of the epidemic for the most affected Northern Italian regions (Piedmont, Lombardy, Veneto and Emilia), giving values of R0 ranging from 2.2 to 3.1. Finally, a theoretical formulation of the infection rate is proposed, introducing a new parameter, the infection length, characteristic of the disease. Keywords: approximate solution; infectious lenght; SIR model. Introduction A sudden increase of cases of Covid-19 diseases originated by the new coronavirus SARS-CoV-2 struck Northern Italy and Lombardy in particular,inlate February 2020 (Distante et al. 2020; Ital- ian National Institute of Health (ISS); Italian Ministry of Health). The rapid growth of many severe illnesses leads to a dramatic pressure on the hospitals, particularly in the intensive care units. -

Dante's Political Life
Bibliotheca Dantesca: Journal of Dante Studies Volume 3 Article 1 2020 Dante's Political Life Guy P. Raffa University of Texas at Austin, [email protected] Follow this and additional works at: https://repository.upenn.edu/bibdant Part of the Ancient, Medieval, Renaissance and Baroque Art and Architecture Commons, Italian Language and Literature Commons, and the Medieval History Commons Recommended Citation Raffa, Guy P. (2020) "Dante's Political Life," Bibliotheca Dantesca: Journal of Dante Studies: Vol. 3 , Article 1. Available at: https://repository.upenn.edu/bibdant/vol3/iss1/1 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/bibdant/vol3/iss1/1 For more information, please contact [email protected]. Raffa: Dante's Political Life Bibliotheca Dantesca, 3 (2020): 1-25 DANTE’S POLITICAL LIFE GUY P. RAFFA, The University of Texas at Austin The approach of the seven-hundredth anniversary of Dante’s death is a propi- tious time to recall the events that drove him from his native Florence and marked his life in various Italian cities before he found his final refuge in Ra- venna, where he died and was buried in 1321. Drawing on early chronicles and biographies, modern historical research and biographical criticism, and the poet’s own writings, I construct this narrative of “Dante’s Political Life” for the milestone commemoration of his death. The poet’s politically-motivated exile, this biographical essay shows, was destined to become one of the world’s most fortunate misfortunes. Keywords: Dante, Exile, Florence, Biography The proliferation of biographical and historical scholarship on Dante in recent years, after a relative paucity of such work through much of the twentieth century, prompted a welcome cluster of re- flections on this critical genre in a recent volume of Dante Studies. -

The North-South Divide in Italy: Reality Or Perception?
CORE Metadata, citation and similar papers at core.ac.uk EUROPEAN SPATIAL RESEARCH AND POLICY Volume 25 2018 Number 1 http://dx.doi.org/10.18778/1231-1952.25.1.03 Dario MUSOLINO∗ THE NORTH-SOUTH DIVIDE IN ITALY: REALITY OR PERCEPTION? Abstract. Although the literature about the objective socio-economic characteristics of the Italian North- South divide is wide and exhaustive, the question of how it is perceived is much less investigated and studied. Moreover, the consistency between the reality and the perception of the North-South divide is completely unexplored. The paper presents and discusses some relevant analyses on this issue, using the findings of a research study on the stated locational preferences of entrepreneurs in Italy. Its ultimate aim, therefore, is to suggest a new approach to the analysis of the macro-regional development gaps. What emerges from these analyses is that the perception of the North-South divide is not consistent with its objective economic characteristics. One of these inconsistencies concerns the width of the ‘per- ception gap’, which is bigger than the ‘reality gap’. Another inconsistency concerns how entrepreneurs perceive in their mental maps regions and provinces in Northern and Southern Italy. The impression is that Italian entrepreneurs have a stereotyped, much too negative, image of Southern Italy, almost a ‘wall in the head’, as also can be observed in the German case (with respect to the East-West divide). Keywords: North-South divide, stated locational preferences, perception, image. 1. INTRODUCTION The North-South divide1 is probably the most known and most persistent charac- teristic of the Italian economic geography. -

ITINERARY: Milan, Bologna, Tuscany, Rome
ITINERARY: Milan, Bologna, Tuscany, Rome 13 May - 25 May 2018 UVU Culinary Arts Institute Day 1, May 13: USA / Milan, Italy Meals: D Arrival: Welcome to italy, one of the most famous travel destinations in the world! Prepare for an incredible journey as you experience everything Italy has to offer from the treasured cultural and historic sites to the world famous cuisine -arguably the most influential component of italian culture. From the aromatic white truffles of Montone to the rich seafood of the Cinque Terre coast, each province and region of Italy offers culinary treats for the most experienced of palates. Our journey will begin in Milan, one of the most prominent Italian cities located at the northernmost tip of Italy. Afternoon: The group will arrive at the Milan MXP (Milan Malpensa International Airport), where upon clearing customs and immigration your private driver and guide will greet the group and assist with the transfer to your hotel. Your group will be staying at the Kilma Hotel in Milan, a stylish modern 4-star hotel located 30 minutes from the airport. Upon arrival, the group will check-in to the hotel and prepare for the start of the experience. Evening: Once the entire group has arrived, we will have a delicious welcome dinner at a local favorite restaurant where we will sample some of the culinary flavors of Northern italy. After a delicious meal the group will retire for the evening. Kilma Hotel Milan Fiere Address: Via Privata Venezia Giulia, 8, 20157 Milano MI, Italy Phone: +39 02 455 0461 Day 2, May 14: Milan, Italy Meals: B, D Breakfast: Before setting out to explore the city of Milan, the group will have a delicious breakfast at the hotel where the group will be able to sample a variety of cheeses, charcuterie, and other traditional breakfast items. -

Emilia-Romagna & San Marino
© Lonely Planet 425 Emilia-Romagna & San Marino Emilia-Romagna has long been overlooked as little more than a stepping stone between the Veneto and Tuscany. But take time to explore this underrated region and you’ll discover an area rich in art and culture, an area of mouthwatering food and robust wine, of cosmopolitan EMILIA-ROMAGNA & SAN MARINO resorts and quiet backwaters. Much of its medieval architecture dates to the Renaissance, when a handful of power ful families set up court here: the Farnese in Parma and Piacenza, the Este in Ferrara and Modena, and the Bentivoglio in Bologna. The regional capital, Bologna, is one of Italy’s unsung joys. A foodie city with a hedonistic approach to life, it’s home to Europe’s oldest university and a stunning medieval centre. A short hop to the northwest, Modena boasts a superb Romanesque cathedral and a hint of the gourmet delights that await in Parma, the city that gifted the world prosciutto crudo (cured ham, popularly known as Parma ham) and parmigiano reggiano (Parmesan). In the countryside to the south, castles pepper hilltops as flat plains give way to the Apennine peaks. Ferrara and Ravenna are the highlights of Romagna (the eastern half of Emilia-Romagna). Both are within easy distance of Bologna and both merit a visit – Ferrara for its beautiful Renaissance centre, Ravenna for its sensational Byzantine mosaics. If, after all that high culture, you need a break, head to Rimini where the crowded beaches and cutting-edge clubs promise more earthy pleasures, or San Marino where armies of day-trippers enjoy vast views. -

The Ghibelline Globalists of the Techno-Structure: on the Current Destinies of Empire and Church
Afterword The Ghibelline Globalists of the Techno-Structure: On the Current Destinies of Empire and Church For the past fifty years, the definitive establishment of the great Asian-Amer- ican-European federation and its unchallenged domination over scattered leftovers of inassimilable barbarousness, in Oceania or in Central Africa, had accustomed all peoples, presently clustered into provinces, to the bliss of a uni- versal, and thenceforth imperturbable, peace. No fewer than one hundred and fifty years of wars were needed to achieve this marvelous development […]. Contrary to public proclamations, it wasn’t a vast democratic republic that emerged from the aggregation. Such an eruption of pride could not but raise a new throne, the highest, the strongest, the most radiant there ever was. Gabriel Tarde, Fragment d’histoire future (1896)1 ean Stone’s New World Order (NWO) tells the story of a “Deep State,” of an extraneous apparatus within the American Federation. This foreign entity, which acts in inconspicuous ways, i.e., through Sextremely exclusive lodges and clubs, appears to be bent on taking over the wholesome strata of America, her exceptional manpower and resourc- es, and harnessing them to a vast design of centralized, planetary domina- tion. This “extraneous body” is typically an oligarchic mindset of unmis- takable British make. Professedly “democratic” and “Liberal,” this English drive is, in fact, ferociously elitist and exploitative. To date, it represents the most sophisticated conception of imperial management. Technical- ly, it uses finance and commerce as its consuetudinary instruments of rent- and resource-extraction; politically, it keeps public opinion “in flux” by playing (i.e., scripting) both sides of the electoral spectrum (Left vs. -
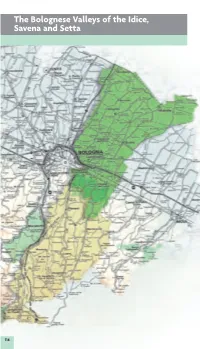
The Bolognese Valleys of the Idice, Savena and Setta
3_ eo_gb 0 008 3: 0 ag a The Bolognese Valleys of the Idice, Savena and Setta 114 _ dce_gb 0 008 3: 9 ag a 5 The Rivers the Futa state highway SS 65 and the road The valleys of the tributaries to the right of along the valley-bottom, which continues as the Reno punctuate the central area of the far as the Lake of Castel dell’Alpi, passing the Bolognese Apennines in a truly surprising majestic Gorges of Scascoli. Along the river, variety of colours and landscapes. They are there are numerous mills, some of which can the Idice, Savena and Setta Rivers, of which be visited, constructed over the centuries. only the Idice continues its course onto the Before entering the plains, the Savena cros- plains, as far as the Park of the Po Delta. ses the Regional Park of Bolognese Gypsums and Abbadessa Gullies, which is also crossed The Idice by the River Idice. The Idice starts on Monte Oggioli, near the Raticosa Pass, and is the largest of the rivers in these valleys. Interesting from a geologi- cal and naturalistic point of view, its valley offers many reasons for a visit. Particularly beautiful is the stretch of river where it joins the Zena Valley: this is where the Canale dei Mulini (mills) branches off, continuing alon- gside it until it reaches the plains, in the ter- ritory of San Lazzaro di Savena. Flowing through the Valleys of Campotto, the Idice finally joins the Reno. Here an interesting system of manmade basins stop the Reno’s water flowing into the Idice’s bed in dry periods. -

Lawyers in the Florence Consular District
Lawyers in the Florence Consular District (The Florence district contains the regions of Emilia-Romagna and Tuscany) Emilia-Romagna Region Disclaimer: The U.S. Consulate General in Florence assumes no responsibility or liability for the professional ability, reputation or the quality of services provided by the persons or firms listed. Inclusion on this list is in no way an endorsement by the Department of State or the U.S. Consulate General. Names are listed alphabetically within each region and the order in which they appear has no other significance. The information on the list regarding professional credentials, areas of expertise and language ability is provided directly by the lawyers. The U.S. Consulate General is not in a position to vouch for such information. You may receive additional information about the individuals by contacting the local bar association or the local licensing authorities. City of Bologna Attorneys Alessandro ALBICINI - Via Marconi 3, 40122 Bologna. Tel: 051/228222-227552. Fax: 051/273323. E- mail: [email protected]. Born 1960. Degree in Jurisprudence. Practice: Commercial law, Industrial, Corportate. Languages: English and French. U.S. correspondents: Kelley Drye & Warren, 101 Park Avenue, New York, NY 10178, Gordon Altman Butowski, 114 West 47th Street, New York, NY 10036- 1510. Luigi BELVEDERI – Via degli Agresti 2, 40123 Bologna. Tel: 051/272600. Fax: 051/271506. E-mail: [email protected]. Born in 1950. Degree in Jurisprudence. Practice: Freelance international attorney since 1978. Languages: English and Italian. Also has office in Milan Via Bigli 2, 20121 Milan Cell: 02780031 Fax: 02780065 Antonio CAPPUCCIO – Piazza Tribunali 6, 40124 Bologna.