Acetylome of Acinetobacter Baumannii SK17 Reveals a Highly-Conserved Modification of Histone-Like Protein HU
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Breast Pathology
30A ANNUAL MEETING ABSTRACTS Conclusions: Our data further demonstrate the complex genetics associated with Results: Patterns identified included: conventional (165 cases, range 10-100%), leiomyosarcomas. As a class, tumor suppressor genes were the most commonly hypocellular/hyalinized (46 cases, range 5-80%), staghorn vessels (35 cases, range mutated loci, including TP53, mutated in 36% of cases (9/25). The loss of key tumor 5-30%), myxoid (27 cases, range 5-40%), keloid (24 cases, range 5-50%), nodular suppressor genes is consistent with our observations that CNV was present in 85% of fasciitis-like (15 cases, range 5-40%), and hypercellular (6 cases, range 5-20%). Tumors leiomyosarcomas. While no common therapeutically targetable gene was identified with keloid areas, as well as those with prominent staghorn blood vessels, mimicked across these cases, future large cohort sequencing studies of leiomyosarcomas may entities such as solitary fibrous tumor. Those cases with nodular fasciitis-like areas raised provide a means for the molecular classification of these tumors and identify new the possibility of nodular fasciitis and reactive processes. Hypercellular foci led to the treatment paradigms. consideration of spindle cell sarcoma, while the differential diagnosis of hypocellular/ hyalinized areas was broad. By site, the greatest variation of patterns was observed in 107 Identification of a Novel FN1-FGFR1 Genetic Fusion as a Frequent intra-abdominal lesions, and men showed more morphologic variability than females. Event in Phosphaturic Mesenchymal Tumor Adults (>18 years) exhibited more histologic diversity than adolescent and pediatric patients (< 18 years). Chang-Tsu Yuan, Yung-Ming Jeng, Sheng-Yao Su, Cher-Wei Liang, Chung-Yen Lin, Conclusions: The morphologic spectrum of desmoid-type fibromatosis is diverse and Shu-Hwa Chen, Cheng-Han Lee, Jodi Carter, Chen-Tu Wu, Andrew Folpe, Jen-Chieh often underappreciated. -

THE OHIO STATE UNIVERSITY Dean's List AUTUMN SEMESTER 2016 Australia Data As of January 19, 2017 Sorted by Zip Code, City and Last Name
THE OHIO STATE UNIVERSITY Dean's List AUTUMN SEMESTER 2016 Australia Data as of January 19, 2017 Sorted by Zip Code, City and Last Name Student Name (Last, First, Middle) City State Zip Bailey, Meg Elizabeth Merewether 2291 Caudle, Emily May Canberra 2609 Davis, Sarah Kate Canberra 2615 Thek, Hannah Louise Surrey Hills 3127 Engel, Rachel Olivia Glen Iris 3146 Taig, Darcy Lachlan Melbourne 3166 THE OHIO STATE UNIVERSITY Enrollment Services - Analysis and Reporting January 19, 2017 Page 1 of 119 Contact: [email protected] THE OHIO STATE UNIVERSITY Dean's List AUTUMN SEMESTER 2016 Bangladesh Data as of January 19, 2017 Sorted by Zip Code, City and Last Name Student Name (Last, First, Middle) City State Zip Bari, Rizvi Dhaka 1215 THE OHIO STATE UNIVERSITY Enrollment Services - Analysis and Reporting January 19, 2017 Page 2 of 119 Contact: [email protected] THE OHIO STATE UNIVERSITY Dean's List AUTUMN SEMESTER 2016 Brazil Data as of January 19, 2017 Sorted by Zip Code, City and Last Name Student Name (Last, First, Middle) City State Zip Rodrigues Franklin, Ana Beatriz Rio de Janeiro 22241 Gomes Pereira Painhas, Henrique Curitiba 80240 Sprintzin, Leonardo Curitiba 80240 Missell, Daniel Caxias do Sul 95020 THE OHIO STATE UNIVERSITY Enrollment Services - Analysis and Reporting January 19, 2017 Page 3 of 119 Contact: [email protected] THE OHIO STATE UNIVERSITY Dean's List AUTUMN SEMESTER 2016 Canada Data as of January 19, 2017 Sorted by Zip Code, City and Last Name Student Name (Last, First, Middle) City State Zip Lu, George Shizhou Vancouver -
Contents More Information
Cambridge University Press 978-1-107-02077-1 — The Cambridge History of China Edited by Albert E. Dien , Keith N. Knapp Table of Contents More Information CONTENTS List of Figures and Tables page x List of Maps xiii Preface xv Six Dynasties Chronology xviii Introduction 1 part 1 history 25 1 Wei 27 by R AFE DE C RESPIGNY Prologue: The Fall of Han (189) 27 Civil War and the Rise of Cao Cao (190–200) 28 Development of a State (200–208) 32 The Limits of Expansion (208–217) 35 From Kingdom to Empire (216–220) 39 Cao Pi and Cao Rui (220–239) 42 Cao Shuang, Sima Yi, and the Fall of Wei (239–265) 46 2 Wu 50 by R AFE DE C RESPIGNY Sun Jian (c.155–191) and Sun Ce (175–200) 50 Sun Quan and the Kingdom of Wu (200–222) 52 The Empire of Sun Quan (222–252) 57 The Succession to Sun Quan and the Fall of Wu (252–280) 61 3 Shu-Han 66 by J. M ICHAEL F ARMER © in this web service Cambridge University Press www.cambridge.org Cambridge University Press 978-1-107-02077-1 — The Cambridge History of China Edited by Albert E. Dien , Keith N. Knapp Table of Contents More Information iv contents The Shu Region in the Late Han 66 The Reign of Liu Yan and Liu Zhang 67 Liu Bei’s Conquest of Yi Province 68 The Reign of Liu Bei (214–223) 70 The Reign of Liu Shan (223–263) 73 4 Western Jin 79 by D AMIEN C HAUSSENDE The Prehistory of the Jin: The Rise of the Sima Clan under the Wei 79 The Reign of Emperor Wu (266–290) 84 The Disturbances of the Eight Princes and the Fall of the Western Jin 92 5 Eastern Jin 96 by C HARLES H OLCOMBE The Founding of the Eastern Jin Dynasty (317–420) 96 Émigrés and Natives 98 Wang Dun’s Rebellion 103 Great-Family Politics 106 Huan Wen 109 The Battle of the Fei River 112 The End of the Eastern Jin 114 An Evaluation 117 6 The Sixteen Kingdoms 119 by C HARLES H OLCOMBE The Emerging Threat 119 The Roads to the Fei River 125 After the Fei River Encounter 137 7 Cheng-Han State 145 by T ERRY F. -

S. No. Name Law Practice 1 A.Revi Shanker S/O K.Annamalai
Annual Election for Council 2021 Category: Middle Nomination Date: 12 October 2020 S. No. Name Law Practice 1 A.Revi Shanker s/o K.Annamalai ARShanker Law Chambers 2 Aaron Kok Ther Chien Holman Fenwick Willan Singapore LLP 3 Aaron Lee Teck Chye Allen & Gledhill LLP 4 Abdul Aziz Bin Abdul Rashid I.R.B. Law LLP 5 Abdul Rahman Bin Mohd Hanipah Abdul Rahman Law Corporation 6 Abdul Wahab Bin Saul Hamid A.W. Law LLC 7 Adam Muneer Yusoff Maniam Drew & Napier LLC 8 Adriel Aloysius Chia Teck Yew Ashurst LLP 9 Ahn Mi Mi Focus Law Asia LLC 10 Aileen Oh Ai Li Fortis Law Corporation 11 Akesh Abhilash Harry Elias Partnership LLP 12 Akshay Kothari Anderson Mori & Tomotsune (Singapore) LLP 13 Alcina Lynn Chew Aiping Tan Kok Quan Partnership 14 Aleksandar Anatoliev Georgiev Rajah & Tann Singapore LLP 15 Alex Yeo Sheng Chye Niru & Co LLC 16 Alexandra Simonetta Jones Gibson, Dunn & Crutcher LLP 17 Allister Brendan Tan Yu Kuan Drew & Napier LLC 18 Alvin Chan Tuan-Tseng (Alvin Zeng Farallon Law Corporation Chuansheng) 19 Alvin Ong Chee Keong (Wang Resource Law LLC Zhiqiang) 20 Amanda Koh Jia Yi Eldan Law LLP 21 Amarjit Singh s/o Hari Singh Amarjit Sidhu Law Practice 22 Amira Nabila Budiyano Gateway Law Corporation 23 Amos Wee Choong Wei DC Law LLC 24 Anand George BR Law Corporation 25 Andre Darius Jumabhoy Peter Low & Choo LLC 26 Ang Ann Liang (Hong Anliang) Allen & Gledhill LLP 27 Ang Chong Yi (Hong Chongyi) RHTLaw Asia LLP 28 Ang Hui Lin, Cheryl Anne (Hong ADTlaw LLC Huilin, Cheryl Anne) 29 Ang Kaili Virtus Law LLP 30 Ang Kim Hock Wong & Leow LLC 31 Ang -
Governing China, 150-1850
Gove rni ng Chi na 15 0–1850 John W. Dardess GOVERNING CHINA 150–1850 GOVERNING CHINA 150–1850 JOHN W. DARDESS Hackett Publishing Company, Inc. Indianapolis/Cambridge Copyright © 2010 by Hackett Publishing Company, Inc. All rights reserved Printed in the United States of America 14 13 12 11 10 1 2 3 4 5 6 7 For further information, please address Hackett Publishing Company, Inc. P.O. Box 44937 Indianapolis, Indiana 46244-0937 www.hackettpublishing.com Cover design by Abigail Coyle Text design by Mary Vasquez Maps by William Nelson Composition by Cohographics Printed at Sheridan Books, Inc. Library of Congress Cataloging-in-Publication Data Dardess, John W., 1937– Governing China : 150–1850 / John W. Dardess. p. cm. Includes bibliographical references and index. ISBN 978-1-60384-311-9 (pbk.) — ISBN 978-1-60384-312-6 (cloth) 1. China—Politics and government. 2. China—Social conditions. 3. China—History—Han dynasty, 202 B.C.–220 A.D. 4. China—History— Qing dynasty, 1644–1912. 5. Political culture—China—History. 6. Social institutions—China—History. 7. Education—China—History. I. Title. DS740.2.D37 2010 951—dc22 2010015241 The paper used in this publication meets the minimum requirements of American National Standard for Information Sciences— Permanence of paper for Printed Library Materials, ANSI z39.48–1984. CONTENTS Preface vii Introduction: Comparing China in 150 and China in 1850 x Timelines xxiii Maps xxvii PART 1. FROM FRAGMENTATION TO REUNIFICATION, 150–589 1 The Unraveling of the Later Han, 150–220 3 The Three Kingdoms, 221–264 5 The Western Jin, 266–311 6 A Fractured Age, 311–450 8 Unity in the North: The Northern Wei, 398–534 12 Not by Blood Alone: Steps to Reunification, 534–589 16 PART 2. -

China 7344 Ng and Wang / MIRRORING the PAST / Sheet
Tseng 2005.6.21 13:11 7344 Ng and Wang / MIRRORING THE PAST / sheet 1 of 332 MIRRORING THE PAST MIRRORING THE PAST Tseng 2005.6.21 13:11 7344 Ng and Wang / MIRRORING THE PAST / sheet 2 of 332 3 of 332 MIRRORINGMIRRORING THE THE PAST PAST The Writing and Use of History in Imperial China 7344 Ng and Wang / MIRRORING THE PAST / sheet On-cho Ng and Q. Edward Wang University of Hawai‘i Press Honolulu Tseng 2005.6.21 13:11 4 of 332 © 2005 University of Hawai‘i Press All rights reserved Printed in the United States of America 100908070605 654321 Library of Congress Cataloging-in-Publication Data 7344 Ng and Wang / MIRRORING THE PAST / sheet Ng, On Cho. Mirroring the past : the writing and use of history in imperial China / On-cho Ng and Q. Edward Wang. p. cm. Includes bibliographical references. ISBN-13: 978-0-8248-2913-1 (hardcover : alk. paper) ISBN-10: 0-8248-2913-1 (hardcover : alk. paper) 1. China—Historiography. 2. Historiography—China— History. I. Title: Writing and use of history in imperial China. II. Wang, Qingjia. III. Title. DS734.7.N4 2005 907'.2'051—dc22 2005008008 University of Hawai‘i Press books are printed on acid-free paper and meet the guidelines for permanence and durability of the Council on Library Resources. Designed by University of Hawai‘i Press production staff Printed by Integrated Book Technology Tseng 2005.6.21 13:11 5 of 332 Contents 7344 Ng and Wang / MIRRORING THE PAST / sheet Prologue vii Chapter 1 The Age of Confucius: The Genesis of History 1 Chapter 2 From the Warring States Period to the Han: The Formation -

The Three Kingdoms, the Wei, Jin and the Southern and Northern Dynasties
Chapter 4 Many Chinas: The Three Kingdoms, the Wei, Jin and the Southern and Northern Dynasties (220-589 AD) Key ideas: This chapter is a brief investigation of a confused, momentous and neglected period. It addresses the main issues, namely the southward shift of the centre of gravity of China in terms both of population and culture and the introduction of Buddhism. In addition, less prominent developments that contributed to the reshaping of China are discussed, such as the formation of medieval aristocracy, the increasing seaward orientation, as well as the formation of southern lifestyle familiar with boats, waterways rice fields and lush vegetation. The period from the fall of the Han to the reunification of China proper by the Sui 随 dynasty is one of the least understood periods of Chinese history. Often dismissed as a period of political disunity it appears merely as a gap between the early and the middle empire. The period is referred by various designations, none of which is particularly satisfactory. It is characterized as a period of political disunity (implying that cultural coherence survived); or also simply as the Six Dynasties (referring to the six states traditional Chinese historians canonized as the legitimate inheritors of the mandate of heaven among the many states of the period); or subdivided into the periods of the Three Kingdoms (which arose from the fallen Han empire) or the Wei (the most powerful of these three states), the Jin (which briefly unified large parts of China), and the Southern and Northern Dynasties (a period of unstable states and an established division between North and South) – an enumeration that is quite short and handy in Chinese: 魏晋南北朝 Wei-Jin-Nanbei chao. -

Modern Pathology Abstracts Index of Abstract Authors
VOLUME 33 | SUPPLEMENT 2 | MARCH 2020 MODERN PATHOLOGY ABSTRACTS INDEX OF ABSTRACT AUTHORS LOS ANGELES CONVENTION CENTER FEBRUARY 29-MARCH 5, 2020 LOS ANGELES, CALIFORNIA 2020 ABSTRACTS | PLATFORM & POSTER PRESENTATIONS EDUCATION COMMITTEE Jason L. Hornick, Chair William C. Faquin Rhonda K. Yantiss, Chair, Abstract Review Board Yuri Fedoriw and Assignment Committee Karen Fritchie Laura W. Lamps, Chair, CME Subcommittee Lakshmi Priya Kunju Anna Marie Mulligan Steven D. Billings, Interactive Microscopy Subcommittee Rish K. Pai Raja R. Seethala, Short Course Coordinator David Papke, Pathologist-in-Training Ilan Weinreb, Subcommittee for Unique Live Course Offerings Vinita Parkash David B. Kaminsky (Ex-Officio) Carlos Parra-Herran Anil V. Parwani Zubair Baloch Rajiv M. Patel Daniel Brat Deepa T. Patil Ashley M. Cimino-Mathews Lynette M. Sholl James R. Cook Nicholas A. Zoumberos, Pathologist-in-Training Sarah Dry ABSTRACT REVIEW BOARD Benjamin Adam Billie Fyfe-Kirschner Michael Lee Natasha Rekhtman Narasimhan Agaram Giovanna Giannico Cheng-Han Lee Jordan Reynolds Rouba Ali-Fehmi Anthony Gill Madelyn Lew Michael Rivera Ghassan Allo Paula Ginter Zaibo Li Andres Roma Isabel Alvarado-Cabrero Tamara Giorgadze Faqian Li Avi Rosenberg Catalina Amador Purva Gopal Ying Li Esther Rossi Roberto Barrios Anuradha Gopalan Haiyan Liu Peter Sadow Rohit Bhargava Abha Goyal Xiuli Liu Steven Salvatore Jennifer Boland Rondell Graham Yen-Chun Liu Souzan Sanati Alain Borczuk Alejandro Gru Lesley Lomo Anjali Saqi Elena Brachtel Nilesh Gupta Tamara Lotan Jeanne Shen -

The Limited Partnership in China
THE LIMITED PARTNERSHIP IN CHINA: AN EVALUATION LIN LIN LL.B. (Hons), GDUFS LL.M., NUS A THESIS SUBMITTED FOR THE DEGREE OF DOCTOR OF PHILOSOPHY FACULTY OF LAW NATIONAL UNIVERSITY OF SINGAPORE 2010 ACKNOWLEDGEMENT First and foremost, I would like to thank National University of Singapore (NUS) for granting me the President‘s Graduate Fellowship and the Research Scholarship to write this thesis. During my doctoral study, I was generously supported by NUS Faculty of Law‘s Research and Conference Fund to present and publish several academic articles. My deepest thanks go to my supervisor, Prof. Yeo Hwee Ying. I would like to thank her for her supervision, guidance, encouragement and support throughout my PhD study. Her constructive feedback and insightful suggestions at every stage have been significantly useful in shaping this thesis up to completion. Without her, a project of this magnitude could not have been brought to a successful completion. In addition to receiving her valuable guidance on this thesis, I had the good fortune of co-writing an article with her. I have learned immensely from this experience. Her conscientiousness, wisdom, kindness and infinite patience have enriched my growth as a legal scholar. I would like to thank Prof. Hans Tjio and Prof. Wee Meng Seng for reviewing my research proposal and for their comments during my doctoral candidate qualifying examination. I also thank their inspiration and encouragement during my graduate study. I would like to express my deep thanks to Prof. Dan W. Puchniak and Prof. 0HWang Jiangyu who gave me extremely constructive and i detailed suggestions during and after my PhD oral defense. -

Routledge Handbook of Imperial Chinese History the Eastern
This article was downloaded by: 10.3.98.104 On: 28 Sep 2021 Access details: subscription number Publisher: Routledge Informa Ltd Registered in England and Wales Registered Number: 1072954 Registered office: 5 Howick Place, London SW1P 1WG, UK Routledge Handbook of Imperial Chinese History Victor Cunrui Xiong, Kenneth J. Hammond The Eastern Han Publication details https://www.routledgehandbooks.com/doi/10.4324/9781315726878-5 Rafe de Crespigny Published online on: 02 Oct 2018 How to cite :- Rafe de Crespigny. 02 Oct 2018, The Eastern Han from: Routledge Handbook of Imperial Chinese History Routledge Accessed on: 28 Sep 2021 https://www.routledgehandbooks.com/doi/10.4324/9781315726878-5 PLEASE SCROLL DOWN FOR DOCUMENT Full terms and conditions of use: https://www.routledgehandbooks.com/legal-notices/terms This Document PDF may be used for research, teaching and private study purposes. Any substantial or systematic reproductions, re-distribution, re-selling, loan or sub-licensing, systematic supply or distribution in any form to anyone is expressly forbidden. The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The publisher shall not be liable for an loss, actions, claims, proceedings, demand or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material. 3 THE EASTERN HAN Rafe de Crespigny Foundation and the first rulers 23–88 ce Under the regime of Wang Mang 王莽, the former imperial Liu 劉 clan had lost many of its privileges, and in the winter of 22 ce, members of the family in Nanyang 南陽 rose in rebel- lion. -

The Essence of Governance: the Development of Public Administration in China
THE ESSENCE OF GOVERNANCE: THE DEVELOPMENT OF PUBLIC ADMINISTRATION IN CHINA Yongfei Zhao A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of MASTER OF PUBLIC ADMINISTRATION December 2005 Committee: Dr. D. S. Chauhan, Advisor Dr. John Hoag Dr. Shannon Orr © 2005 Yongfei Zhao All Rights Reserved iii ABSTRACT Dr. D. S. Chauhan, Advisor The government of China has made concerted efforts to modernize the society by undertaking major administrative reforms and by professionalizing public service education since 1999. In addition to various administrative reform programs initiated by Deng Xiaoping and implemented by Jiang Zemin and Hu Jintao, the inauguration of Master of Public Administration degree programs has provided the real meaning to those administrative reforms in enhancing the management capacity of government employees. All these efforts are directed toward establishing public administration as a discipline. This study provides (1) the historic overview of the development of public administration in China; (2) examines the administrative structures and reforms undertaken by the Chinese government to modernize administrative systems; (3) analyzes the establishment of MPA degree programs to develop professional public service; and (4) assesses the impact of globalization on political, administrative, and economic institutions in Chinese society. Throughout the study, a special attention has been paid to the examination of the impact of Chinese culture, values, tradition, and socialist ideology on the governance process. iv To Dr. Chauhan and My Parents v ACKNOWLEDGEMENTS I am deeply indebted to my academic advisor, Professor D. S. Chauhan, for his constant encouragement and help to complete my thesis. -
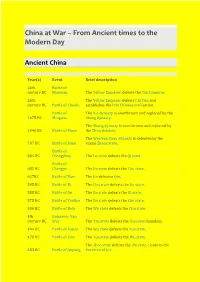
China at War – from Ancient Times to the Modern Day
China at War – From Ancient times to the Modern Day Ancient China Year(s) Event Brief description 26th Battle of century BC Banquan The Yellow Emperor defeats the Yan Emperor. 26th The Yellow Emperor defeats Chi You and century BC Battle of Zhuolu establishes the Han Chinese civilisation. Battle of The Xia dynasty is overthrown and replaced by the 1675 BC Mingtiao Shang dynasty. The Shang dynasty is overthrown and replaced by 1046 BC Battle of Muye the Zhou dynasty. The Western Zhou dynasty is defeated by the 707 BC Battle of Xuge vassal Zheng state. Battle of 684 BC Changshao The Lu state defeats the Qi state Battle of 632 BC Chengpu The Jin state defeats the Chu state. 627BC Battle of Xiao The Jin defeates Qin. 595 BC Battle of Bi The Chu state defeats the Jin state. 588 BC Battle of An The Jin state defeats the Qi state. 575 BC Battle of Yanling The Jin state defeats the Chu state. 506 BC Battle of Boju The Wu state defeats the Chu state. 4th Gojoseon–Yan century BC War The Yan state defeats the Gojoseon kingdom. 494 BC Battle of Fujiao The Wu state defeats the Yue state. 478 BC Battle of Lize The Yue state defeats the Wu state. The Zhao state defeats the Zhi state. Leads to the 453 BC Battle of Jinyang Partition of Jin. 353 BC Battle of Guiling The Qi state defeats the Wei state. 342 BC Battle of Maling The Qi state defeats the Wei state. Battle of 341 BC Guailing 293 BC Battle of Yique The Qin state defeats the Wei and Han states.