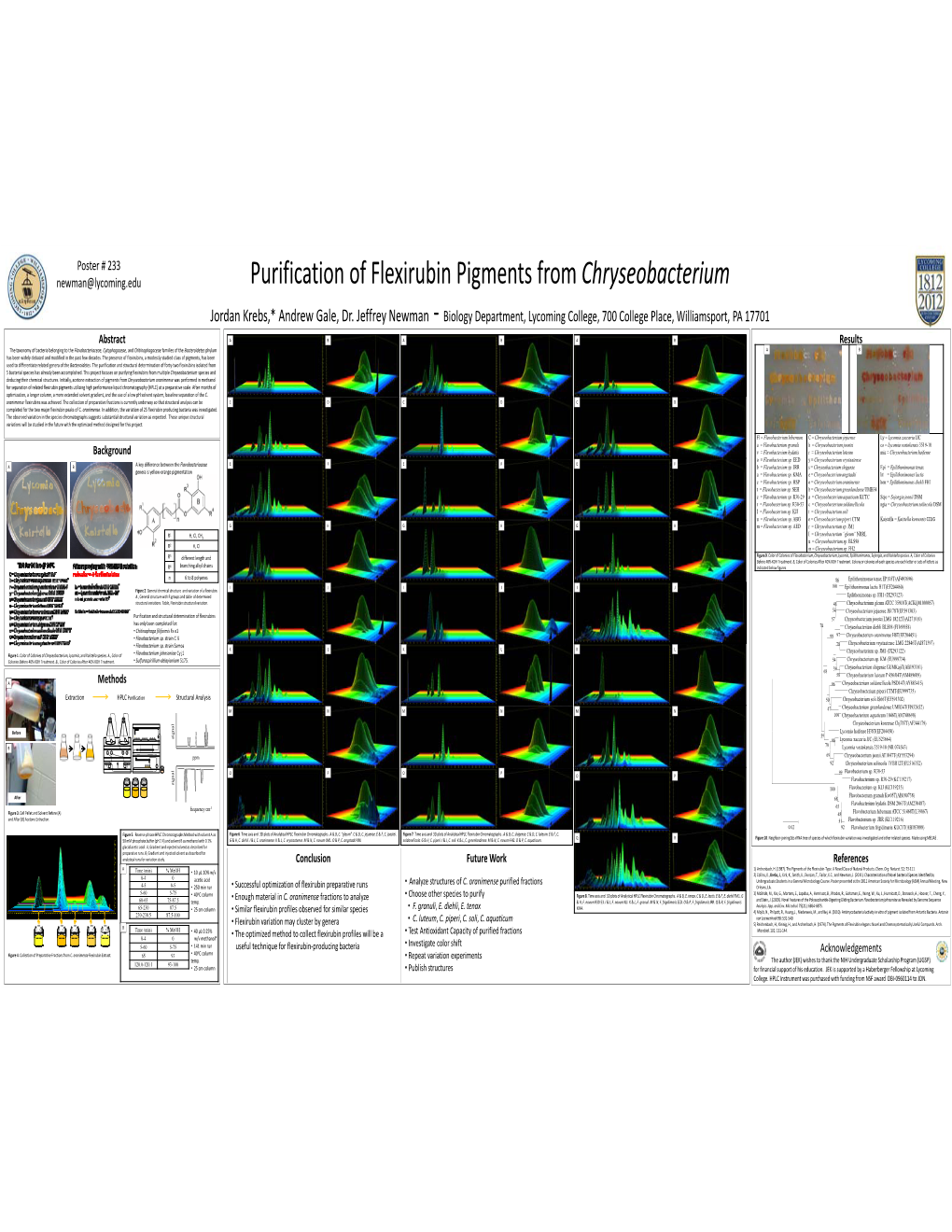
Purification of Flexirubin Pigments from Chryseobacterium Jordan Krebs,* Andrew Gale, Dr
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

High Quality Permanent Draft Genome Sequence of Chryseobacterium Bovis DSM 19482T, Isolated from Raw Cow Milk
Lawrence Berkeley National Laboratory Recent Work Title High quality permanent draft genome sequence of Chryseobacterium bovis DSM 19482T, isolated from raw cow milk. Permalink https://escholarship.org/uc/item/4b48v7v8 Journal Standards in genomic sciences, 12(1) ISSN 1944-3277 Authors Laviad-Shitrit, Sivan Göker, Markus Huntemann, Marcel et al. Publication Date 2017 DOI 10.1186/s40793-017-0242-6 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Laviad-Shitrit et al. Standards in Genomic Sciences (2017) 12:31 DOI 10.1186/s40793-017-0242-6 SHORT GENOME REPORT Open Access High quality permanent draft genome sequence of Chryseobacterium bovis DSM 19482T, isolated from raw cow milk Sivan Laviad-Shitrit1, Markus Göker2, Marcel Huntemann3, Alicia Clum3, Manoj Pillay3, Krishnaveni Palaniappan3, Neha Varghese3, Natalia Mikhailova3, Dimitrios Stamatis3, T. B. K. Reddy3, Chris Daum3, Nicole Shapiro3, Victor Markowitz3, Natalia Ivanova3, Tanja Woyke3, Hans-Peter Klenk4, Nikos C. Kyrpides3 and Malka Halpern1,5* Abstract Chryseobacterium bovis DSM 19482T (Hantsis-Zacharov et al., Int J Syst Evol Microbiol 58:1024-1028, 2008) is a Gram-negative, rod shaped, non-motile, facultative anaerobe, chemoorganotroph bacterium. C. bovis is a member of the Flavobacteriaceae, a family within the phylum Bacteroidetes. It was isolated when psychrotolerant bacterial communities in raw milk and their proteolytic and lipolytic traits were studied. Here we describe the features of this organism, together with the draft genome sequence and annotation. The DNA G + C content is 38.19%. The chromosome length is 3,346,045 bp. It encodes 3236 proteins and 105 RNA genes. The C. bovis genome is part of the Genomic Encyclopedia of Type Strains, Phase I: the one thousand microbial genomes study. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Structural Basis of Mammalian Mucin Processing by the Human Gut O
ARTICLE https://doi.org/10.1038/s41467-020-18696-y OPEN Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila ✉ ✉ Beatriz Trastoy 1,4, Andreas Naegeli2,4, Itxaso Anso 1,4, Jonathan Sjögren 2 & Marcelo E. Guerin 1,3 Akkermansia muciniphila is a mucin-degrading bacterium commonly found in the human gut that promotes a beneficial effect on health, likely based on the regulation of mucus thickness 1234567890():,; and gut barrier integrity, but also on the modulation of the immune system. In this work, we focus in OgpA from A. muciniphila,anO-glycopeptidase that exclusively hydrolyzes the peptide bond N-terminal to serine or threonine residues substituted with an O-glycan. We determine the high-resolution X-ray crystal structures of the unliganded form of OgpA, the complex with the glycodrosocin O-glycopeptide substrate and its product, providing a comprehensive set of snapshots of the enzyme along the catalytic cycle. In combination with O-glycopeptide chemistry, enzyme kinetics, and computational methods we unveil the molecular mechanism of O-glycan recognition and specificity for OgpA. The data also con- tribute to understanding how A. muciniphila processes mucins in the gut, as well as analysis of post-translational O-glycosylation events in proteins. 1 Structural Biology Unit, Center for Cooperative Research in Biosciences (CIC bioGUNE), Basque Research and Technology Alliance (BRTA), Bizkaia Technology Park, Building 801A, 48160 Derio, Spain. 2 Genovis AB, Box 790, 22007 Lund, Sweden. 3 IKERBASQUE, Basque Foundation for Science, 48013 ✉ Bilbao, Spain. 4These authors contributed equally: Beatriz Trastoy, Andreas Naegeli, Itxaso Anso. -

Emerging Flavobacterial Infections in Fish
Journal of Advanced Research (2014) xxx, xxx–xxx Cairo University Journal of Advanced Research REVIEW Emerging flavobacterial infections in fish: A review Thomas P. Loch a, Mohamed Faisal a,b,* a Department of Pathobiology and Diagnostic Investigation, College of Veterinary Medicine, 174 Food Safety and Toxicology Building, Michigan State University, East Lansing, MI 48824, USA b Department of Fisheries and Wildlife, College of Agriculture and Natural Resources, Natural Resources Building, Room 4, Michigan State University, East Lansing, MI 48824, USA ARTICLE INFO ABSTRACT Article history: Flavobacterial diseases in fish are caused by multiple bacterial species within the family Received 12 August 2014 Flavobacteriaceae and are responsible for devastating losses in wild and farmed fish stocks Received in revised form 27 October 2014 around the world. In addition to directly imposing negative economic and ecological effects, Accepted 28 October 2014 flavobacterial disease outbreaks are also notoriously difficult to prevent and control despite Available online xxxx nearly 100 years of scientific research. The emergence of recent reports linking previously uncharacterized flavobacteria to systemic infections and mortality events in fish stocks of Keywords: Europe, South America, Asia, Africa, and North America is also of major concern and has Flavobacterium highlighted some of the difficulties surrounding the diagnosis and chemotherapeutic treatment Chryseobacterium of flavobacterial fish diseases. Herein, we provide a review of the literature that focuses on Fish disease Flavobacterium and Chryseobacterium spp. and emphasizes those associated with fish. Coldwater disease ª 2014 Production and hosting by Elsevier B.V. on behalf of Cairo University. Flavobacteriosis Mohamed Faisal D.V.M., Ph.D., is currently a Thomas P. -

Chryseobacterium Oleae Sp. Nov., an Efficient Plant
Systematic and Applied Microbiology 37 (2014) 342–350 Contents lists available at ScienceDirect Systematic and Applied Microbiology j ournal homepage: www.elsevier.de/syapm Short communication Chryseobacterium oleae sp. nov., an efficient plant growth promoting bacterium in the rooting induction of olive tree (Olea europaea L.) cuttings and emended descriptions of the genus Chryseobacterium, C. daecheongense, C. gambrini, C. gleum, C. joostei, C. jejuense, C. luteum, C. shigense, C. taiwanense, C. ureilyticum and C. vrystaatense a,b,∗∗ a c Maria del Carmen Montero-Calasanz , Markus Göker , Manfred Rohde , a a d e Cathrin Spröer , Peter Schumann , Hans-Jürgen Busse , Michael Schmid , a a,∗ b Hans-Peter Klenk , Brian J. Tindall , Maria Camacho a Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures, Inhoffenstraße 7B, 38124 Braunschweig, Germany b IFAPA-Instituto de Investigación y Formación Agraria y Pesquera, Centro Las Torres-Tomejil, Ctra, Sevilla-Cazalla de la Sierra, Km 12.2, Alcalá del Río, 41200 Sevilla, Spain c HZI – Helmholtz Centre for Infection Research, Inhoffenstraße 7, 38124 Braunschweig, Germany d Institut für Bakteriologie, Mykologie und Hygiene, Veterinärmedizinische Universität, A-1210 Wien, Austria e Research Unit Microbe-Plant Interactions, Helmholtz Zentrum München, Ingolstädter Landstraße 1, 85764 Neuherberg, Germany a r t i c l e i n f o a b s t r a c t T Article history: A novel non-motile, Gram-staining-negative, yellow-pigmented bacterium, designated CT348 , isolated Received 5 November 2013 from the ectorhizosphere of an organic olive tree in Spain and characterised as an efficient plant growth Received in revised form 21 February 2014 promoting bacterium, was investigated to determine its taxonomic status. -

Bacteria Associated with Vascular Wilt of Poplar
Bacteria associated with vascular wilt of poplar Hanna Kwasna ( [email protected] ) Poznan University of Life Sciences: Uniwersytet Przyrodniczy w Poznaniu https://orcid.org/0000-0001- 6135-4126 Wojciech Szewczyk Poznan University of Life Sciences: Uniwersytet Przyrodniczy w Poznaniu Marlena Baranowska Poznan University of Life Sciences: Uniwersytet Przyrodniczy w Poznaniu Jolanta Behnke-Borowczyk Poznan University of Life Sciences: Uniwersytet Przyrodniczy w Poznaniu Research Article Keywords: Bacteria, Pathogens, Plantation, Poplar hybrids, Vascular wilt Posted Date: May 27th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-250846/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/30 Abstract In 2017, the 560-ha area of hybrid poplar plantation in northern Poland showed symptoms of tree decline. Leaves appeared smaller, turned yellow-brown, and were shed prematurely. Twigs and smaller branches died. Bark was sunken and discolored, often loosened and split. Trunks decayed from the base. Phloem and xylem showed brown necrosis. Ten per cent of trees died in 1–2 months. None of these symptoms was typical for known poplar diseases. Bacteria in soil and the necrotic base of poplar trunk were analysed with Illumina sequencing. Soil and wood were colonized by at least 615 and 249 taxa. The majority of bacteria were common to soil and wood. The most common taxa in soil were: Acidobacteria (14.757%), Actinobacteria (14.583%), Proteobacteria (36.872) with Betaproteobacteria (6.516%), Burkholderiales (6.102%), Comamonadaceae (2.786%), and Verrucomicrobia (5.307%).The most common taxa in wood were: Bacteroidetes (22.722%) including Chryseobacterium (5.074%), Flavobacteriales (10.873%), Sphingobacteriales (9.396%) with Pedobacter cryoconitis (7.306%), Proteobacteria (73.785%) with Enterobacteriales (33.247%) including Serratia (15.303%) and Sodalis (6.524%), Pseudomonadales (9.829%) including Pseudomonas (9.017%), Rhizobiales (6.826%), Sphingomonadales (5.646%), and Xanthomonadales (11.194%). -

Epidemiological and Phylogenetic Analysis Reveals Flavobacteriaceae As Potential Ancestral Source of Tigecycline Resistance Gene Tet(X)
ARTICLE https://doi.org/10.1038/s41467-020-18475-9 OPEN Epidemiological and phylogenetic analysis reveals Flavobacteriaceae as potential ancestral source of tigecycline resistance gene tet(X) Rong Zhang 1,5, Ning Dong2,5, Zhangqi Shen3,5, Yu Zeng1, Jiauyue Lu1, Congcong Liu1, Hongwei Zhou1, Yanyan Hu1, Qiaoling Sun1, Qipeng Cheng2,4, Lingbing Shu1, Jiachang Cai1, Edward Wai-Chi Chan4, ✉ ✉ Gongxiang Chen 1 & Sheng Chen 2 1234567890():,; Emergence of tigecycline-resistance tet(X) gene orthologues rendered tigecycline ineffective as last-resort antibiotic. To understand the potential origin and transmission mechanisms of these genes, we survey the prevalence of tet(X) and its orthologues in 2997 clinical E. coli and K. pneumoniae isolates collected nationwide in China with results showing very low pre- valence on these two types of strains, 0.32% and 0%, respectively. Further surveillance of tet (X) orthologues in 3692 different clinical Gram-negative bacterial strains collected during 1994–2019 in hospitals in Zhejiang province, China reveals 106 (2.7%) tet(X)-bearing strains with Flavobacteriaceae being the dominant (97/376, 25.8%) bacteria. In addition, tet(X)s are found to be predominantly located on the chromosomes of Flavobacteriaceae and share similar GC-content as Flavobacteriaceae. It also further evolves into different orthologues and transmits among different species. Data from this work suggest that Flavobacteriaceae could be the potential ancestral source of the tigecycline resistance gene tet(X). 1 Department of Clinical Laboratory, Second Affiliated Hospital of Zhejiang University, School of Medicine, ZhejiangHangzhou, China. 2 Department of Infectious Diseases and Public Health, Jockey Club College of Veterinary Medicine and Life Sciences, City University of Hong Kong, Kowloon, Hong Kong, China. -

Genome-Based Taxonomic Classification Of
ORIGINAL RESEARCH published: 20 December 2016 doi: 10.3389/fmicb.2016.02003 Genome-Based Taxonomic Classification of Bacteroidetes Richard L. Hahnke 1 †, Jan P. Meier-Kolthoff 1 †, Marina García-López 1, Supratim Mukherjee 2, Marcel Huntemann 2, Natalia N. Ivanova 2, Tanja Woyke 2, Nikos C. Kyrpides 2, 3, Hans-Peter Klenk 4 and Markus Göker 1* 1 Department of Microorganisms, Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany, 2 Department of Energy Joint Genome Institute (DOE JGI), Walnut Creek, CA, USA, 3 Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia, 4 School of Biology, Newcastle University, Newcastle upon Tyne, UK The bacterial phylum Bacteroidetes, characterized by a distinct gliding motility, occurs in a broad variety of ecosystems, habitats, life styles, and physiologies. Accordingly, taxonomic classification of the phylum, based on a limited number of features, proved difficult and controversial in the past, for example, when decisions were based on unresolved phylogenetic trees of the 16S rRNA gene sequence. Here we use a large collection of type-strain genomes from Bacteroidetes and closely related phyla for Edited by: assessing their taxonomy based on the principles of phylogenetic classification and Martin G. Klotz, Queens College, City University of trees inferred from genome-scale data. No significant conflict between 16S rRNA gene New York, USA and whole-genome phylogenetic analysis is found, whereas many but not all of the Reviewed by: involved taxa are supported as monophyletic groups, particularly in the genome-scale Eddie Cytryn, trees. Phenotypic and phylogenomic features support the separation of Balneolaceae Agricultural Research Organization, Israel as new phylum Balneolaeota from Rhodothermaeota and of Saprospiraceae as new John Phillip Bowman, class Saprospiria from Chitinophagia. -
Structural and Functional Diversity of the Diazotrophic Community in Xeric Ecosystems: Response to Nitrogen Availability
UNIVERSIDADE DE LISBOA FACULDADE DE CIÊNCIAS DEPARTAMENTO BIOLOGIA VEGETAL Structural and functional diversity of the diazotrophic community in xeric ecosystems: response to nitrogen availability Carolina Cristiano de Almeida Mestrado em Microbiologia Aplicada Dissertação orientada por: Prof. Cristina Maria Nobre Sobral de Vilhena da Cruz Houghton Prof. Rogério Paulo de Andrade Tenreiro 2019 This Dissertation was fully performed at Plant Soil Ecology at cE3c under the direct co-supervision of Prof. Cristina Maria Nobre Sobral de Vilhena da Cruz Houghton Professor Rogério Paulo de Andrade Tenreiro was the internal supervisor designated in the scope of the Master in Applied Microbiology of the Faculty of Sciences of the University of Lisbon ACKNOWLEDGMENTS This dissertation arose from the collaboration between the Plant Soil Ecology group at cE3c and the Laboratory of Microbiology and Biotechnology at BioISI. This work would not be possible without the assistance and commitment of the individuals involved. First, I would like to express my greatest gratefulness to Professor Cristina Cruz, for the opportunity that was given to me. Thank you for the knowledge shared and for always having an enthusiastic perspective which is contagious. To Professor Rogério Tenreiro, thank you for welcoming me, for always pushing me to see further and for helping me to build more critical thinking. Thank you for all the knowledge shared with me, and for the ideas that help my work to go beyond what I thought was possible. I would also like to thank all members of the PSE group for their contribution, especially to Teresa Dias who has made possible this work with the samples used as well as every soil characteristic. -

Identification of Novel Flavobacteria from Michigan and Assessment of Their Impacts on Fish Health
IDENTIFICATION OF NOVEL FLAVOBACTERIA FROM MICHIGAN AND ASSESSMENT OF THEIR IMPACTS ON FISH HEALTH By Thomas P. Loch A DISSERTATION Submitted to Michigan State University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Pathology 2012 1 ABSTRACT IDENTIFICATION OF NOVEL FLAVOBACTERIA FROM MICHIGAN AND ASSESSMENT OF THEIR IMPACTS ON FISH HEALTH By Thomas P. Loch Flavobacteriosis poses a serious threat to wild and propagated fish stocks alike, accounting for more fish mortality in the State of Michigan, USA, and its associated hatcheries than all other pathogens combined. Although this consortium of fish diseases has primarily been attributed to Flavobacterium psychrophilum, F. columnare, and F. branchiophilum, herein I describe a diverse assemblage of Flavobacterium spp. and Chryseobacterium spp. recovered from diseased, as well as apparently healthy wild, feral, and famed fishes of Michigan. Among 254 fish-associated flavobacterial isolates recovered from 21 fish species during 2003-2010, 211 of these isolates were Flavobacterium spp., and 43 were Chryseobacterium spp. according to ribosomal RNA partial gene sequencing and phylogenetic analysis. Both F. psychrophilum and F. columnare were indeed associated with multiple fish epizootics, but the majority of isolates were either most similar to recently described Flavobacterium and Chryseobacterium spp. that have not been reported within North America, or they did not cluster with any described species. Many of these previously uncharacterized flavobacteria were recovered from systemically infected fish that showed overt signs of disease and were highly proteolytic to multiple substrates in protease assays. Polyphasic characterization, which included extensive physiological, morphological, and biochemical analyses, fatty acid profiling, and phylogenetic analyses using Bayesian and neighbor-joining methodologies, confirmed that there were at least eight clusters of isolates that belonged to the genera Chryseobacterium and Flavobacterium, which represented eight novel species. -

TAXONOMY, GROWTH and FOOD SPOILAGE CHARACTERISTICS of a NOVEL Chryseobacterium SPECIES
TAXONOMY, GROWTH AND FOOD SPOILAGE CHARACTERISTICS OF A NOVEL Chryseobacterium SPECIES By Lize Oosthuizen Submitted in fulfilment of the requirements for the degree of Magister Scientiae (Microbiology) In the Department of Microbial, Biochemical and Food Biotechnology Faculty of Natural and Agricultural Sciences University of the Free State Supervisor: Prof. C. J. Hugo Co-supervisors: Dr. G. Charimba, Prof. J. D. Newman, Dr. A. Hitzeroth, Mrs. L. Steyn November 2018 DECLARATION I declare that the dissertation hereby submitted by me for the M. Sc. Degree in the Faculty of Natural and Agricultural Science at the University of the Free State is my own independent work and has not previously been submitted by me at another university/faculty. I furthermore cede copyright of the dissertation in favour of the University of the Free State. L. Oosthuizen November, 2018 TABLE OF CONTENTS Chapter Title Page TABLE OF CONTENTS i ACKNOWLEDGEMENTS iii LIST OF TABLES iv LIST OF FIGURES vi LIST OF ABBREVIATIONS ix 1 INTRODUCTION 1 2 LITERATURE REVIEW 5 2.1 Introduction 5 2.2 The genus Chryseobacterium 8 2.2.1 History 8 2.2.2 Characteristics 9 2.2.3 Significance of Chryseobacterium species in food 10 2.3 Description of novel Chryseobacterium species using a 14 polyphasic approach 2.3.1 Genotypic methods 15 2.3.2 Phenotypic characterization 21 2.3.3 Chemotaxonomic methods 24 2.4 Growth kinetics of Chryseobacterium species 27 2.4.1 Microbial growth phases 27 2.4.2 Methods for measuring microbial growth 28 2.4.3 Factors influencing microbial growth and food -

Genome-Based Taxonomic Classification of Bacteroidetes
Lawrence Berkeley National Laboratory Recent Work Title Genome-Based Taxonomic Classification of Bacteroidetes. Permalink https://escholarship.org/uc/item/2fs841cf Journal Frontiers in microbiology, 7(DEC) ISSN 1664-302X Authors Hahnke, Richard L Meier-Kolthoff, Jan P García-López, Marina et al. Publication Date 2016 DOI 10.3389/fmicb.2016.02003 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ORIGINAL RESEARCH published: 20 December 2016 doi: 10.3389/fmicb.2016.02003 Genome-Based Taxonomic Classification of Bacteroidetes Richard L. Hahnke 1 †, Jan P. Meier-Kolthoff 1 †, Marina García-López 1, Supratim Mukherjee 2, Marcel Huntemann 2, Natalia N. Ivanova 2, Tanja Woyke 2, Nikos C. Kyrpides 2, 3, Hans-Peter Klenk 4 and Markus Göker 1* 1 Department of Microorganisms, Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany, 2 Department of Energy Joint Genome Institute (DOE JGI), Walnut Creek, CA, USA, 3 Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia, 4 School of Biology, Newcastle University, Newcastle upon Tyne, UK The bacterial phylum Bacteroidetes, characterized by a distinct gliding motility, occurs in a broad variety of ecosystems, habitats, life styles, and physiologies. Accordingly, taxonomic classification of the phylum, based on a limited number of features, proved difficult and controversial in the past, for example, when decisions were based on unresolved phylogenetic trees of the 16S rRNA gene sequence. Here we use a large collection of type-strain genomes from Bacteroidetes and closely related phyla for Edited by: assessing their taxonomy based on the principles of phylogenetic classification and Martin G.