Genetic Diversity Among Juglans Regiaand Prunus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Essential Wholesale & Labs Carrier Oils Chart
Essential Wholesale & Labs Carrier Oils Chart This chart is based off of the virgin, unrefined versions of each carrier where applicable, depending on our website catalog. The information provided may vary depending on the carrier's source and processing and is meant for educational purposes only. Viscosity Absorbtion Comparible Subsitutions Carrier Oil/Butter Color (at room Odor Details/Attributes Rate (Based on Viscosity & Absorbotion Rate) temperature) Description: Stable vegetable butter with a neutral odor. High content of monounsaturated oleic acid and relatively high content of natural antioxidants. Offers good oxidative stability, excellent Almond Butter White to pale yellow Soft Solid Fat Neutral Odor Average cold weather stability, contains occlusive properties, and can act as a moistening agent. Aloe Butter, Illipe Butter Fatty Acid Compositon: Palmitic, Stearic, Oleic, and Linoleic Description: Made from Aloe Vera and Coconut Oil. Can be used as an emollient and contains antioxidant properties. It's high fluidiy gives it good spreadability, and it can quickly hydrate while Aloe Butter White Soft Semi-Solid Fat Neutral Odor Average being both cooling and soothing. Fatty Acid Almond Butter, Illipe Butter Compostion: Linoleic, Oleic, Palmitic, Stearic Description: Made from by combinging Aloe Vera Powder with quality soybean oil to create a Apricot Kernel Oil, Broccoli Seed Oil, Camellia Seed Oil, Evening Aloe Vera Oil Clear, off-white to yellow Free Flowing Liquid Oil Mild musky odor Fast soothing and nourishing carrier oil. Fatty Acid Primrose Oil, Grapeseed Oil, Meadowfoam Seed Oil, Safflower Compostion: Linoleic, Oleic, Palmitic, Stearic Oil, Strawberry Seed Oil Description: This oil is similar in weight to human sebum, making it extremely nouirshing to the skin. -
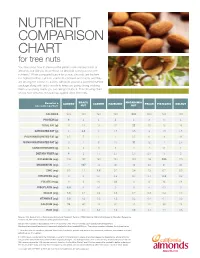
Nutrient Comparison Chart
NUTRIENT COMPARISON CHART for tree nuts You may know how to measure the perfect one-ounce portion of almonds, but did you know those 23 almonds come packed with nutrients? When compared ounce for ounce, almonds are the tree nut highest in fiber, calcium, vitamin E, riboflavin and niacin, and they are among the lowest in calories. Almonds provide a powerful nutrient package along with tasty crunch to keep you going strong, making them a satisfying snack you can feel good about. The following chart shows how almonds measure up against other tree nuts. BRAZIL MACADAMIA Based on a ALMOND CASHEW HAZELNUT PECAN PISTACHIO WALNUT one-ounce portion1 NUT NUT CALORIES 1602 190 160 180 200 200 160 190 PROTEIN (g) 6 4 4 4 2 3 6 4 TOTAL FAT (g) 14 19 13 17 22 20 13 19 SATURATED FAT (g) 1 4.5 3 1.5 3.5 2 1.5 1.5 POLYUNSATURATED FAT (g) 3.5 7 2 2 0.5 6 4 13 MONOUNSATURATED FAT (g) 9 7 8 13 17 12 7 2.5 CARBOHYDRATES (g) 6 3 9 5 4 4 8 4 DIETARY FIBER (g) 4 2 1.5 2.5 2.5 2.5 3 2 POTASSIUM (mg) 208 187 160 193 103 116 285 125 MAGNESIUM (mg) 77 107 74 46 33 34 31 45 ZINC (mg) 0.9 1.2 1.6 0.7 0.4 1.3 0.7 0.9 VITAMIN B6 (mg) 0 0 0.1 0.2 0.1 0.1 0.3 0.2 FOLATE (mcg) 12 6 20 32 3 6 14 28 RIBOFLAVIN (mg) 0.3 0 0.1 0 0 0 0.1 0 NIACIN (mg) 1.0 0.1 0.4 0.5 0.7 0.3 0.4 0.3 VITAMIN E (mg) 7.3 1.6 0.3 4.3 0.2 0.4 0.7 0.2 CALCIUM (mg) 76 45 13 32 20 20 30 28 IRON (mg) 1.1 0.7 1.7 1.3 0.8 0.7 1.1 0.8 Source: U.S. -

Is Peanut Butter Keto-Friendly?
Is Peanut Butter Keto-Friendly? Yes! Peanut butter contains carbohydrates, fat, and protein; the ratios of those macronutrients are what makes it acceptable for a keto diet. Our Once Again peanut butters that are made with one simple ingredient: peanuts, are a healthy addition to your keto diet. Peanuts are in the legume family, which includes a variety of beans. Generally speaking, legumes are not included in keto diets; however, peanuts have a higher fat content than other beans which makes them keto-friendly. Moreover, peanuts have a similar macronutrient distribution when compared to nuts such as almonds. They are high in fat and lower in carbs, and thus make a perfect food for the keto lifestyle. Peanut butter fits nicely into your keto diet as a dip for celery, or add it to smoothies, or experiment with it as an ingredient in salad dressings. Is Almond Butter Keto- Friendly? Yes! Almond butter is a staple for most following a keto lifestyle. Used often as a versatile ingredient in salad dressings, dips, and sauces, and in many other culinary innovations, nut butters in general have the desired macronutrient ration for a keto diet, and almond butter is a particularly good choice because of its fiber and vitamin E content. Most of our almond butters contain one simple ingredient: almonds. The newest addition to our product line is our Blanched Almond Butter, available in both in Extra Creamy and Crunchy varieties. Once Again Blanched Almond Butters contain only one gram of carbohydrate per serving! Blanching almonds involves removing the exterior of the almond, leaving behind a surprisingly nutty-sweet interior, with its lighter texture and color. -

Tea Seed Oil and Health Properties Fatih Seyis1, Emine
Tea Seed Oil and Health Properties Fatih Seyis1, Emine Yurteri1, Aysel Özcan1 1Recep Tayyip Erdoğan University: Faculty of Agriculture and Natural Science, Field Crops Department, Rize/Turkey, e-mail: [email protected] Abstract: Tea Oil has a mild fragrant flavor that goes with anything. It’s not a heavy oil like Olive Oil, but thinner – more like almond oil. If the taste or “oiliness” of olive oil overpowers your food. Along with its mild taste and pleasant tea-like aroma, this oil touts impressive health benefits. Tea seed oil has a high smoke point, contains more monounsaturated fatty acids than olive oil, contains fewer saturated fatty acids than olive oil, contains high levels of Vitamin E, polyphenol antioxidants and both Omegas 3 and 6, but has less Omega 6 and Polyunsaturated Fats than olive oil. Health Benefits of tea seed oil are: it can be applied topically and consumed internally to obtain its health benefits, camellia oil can be used for skin, hair, has anti-cancer effects, effects boost immunity and reduces oxidative stress. Camellia oil is used for a variety of other purposes, for example for cooking, as machinery lubricant, as ingredient in beauty products like night creams, salves, in hair care products and perfumes and is used to coat iron products to prevent rusting. Key words: Tea, seed oil, health 1. Introduction Like other genera of Camellia (from Theaceae family), the tea plant (C.sinensis) produces large oily seeds. In some countries where tea seed oil is abundantly available, it has been accepted as edible oil (Sahari et al., 2004). -

Bioactive Compounds in Nuts and Edible Seeds: Focusing on Brazil Nuts and Baru Almond of the Amazon and Cerrado Brazilian Biomes
Review Article SM Journal of Bioactive Compounds in Nuts and Nutrition and Edible Seeds: Focusing on Brazil Nuts Metabolism and Baru Almond of the Amazon and Cerrado Brazilian Biomes Egea MB1*, Lima DS1, Lodete AR1 and Takeuchi K1,2* 1Science and Technology, Goiano Institute of Education, Brazil 2Faculty of Nutrition, Federal University of Mato Grosso, Brazil Article Information Abstract Received date: Oct 09, 2017 The biodiversity of the Amazon and Cerrado biomes is extremely important for the populations that inhabit Accepted date: Nov 14, 2017 these areas, through the extractive collection of non-timber forest products such as fruits, nuts and edible seeds, which generate income and employment. Brazil nut (Bertholletia excelsa) is native from South America being Published date: Nov 20, 2017 found in the Amazon biome and baru almond (Dipteryx alata Vog.) is native from the Cerrado biome; these are part of the group of oleaginous that can be classified as true nuts and edible seeds, respectively. Both *Corresponding author are important sources of micronutrients that have been associated with several benefits to human health due to the presence of high levels of biologically active compounds such as minerals and vitamins. Minerals act Egea MB, Science and Technology, mostly as cofactors in various reactions, selenium has high availability in Brazil nuts and from selenocysteine Goiano Institute of Education, Brazil, and its enzymes, it exerts functions in the human body as an antioxidant, regulator of thyroid hormones and Tel: +55 64 36205636; protection of cardiovascular diseases. Among vitamins, tocopherol is a precursor to vitamin E, present in both Brazil nut and baru almond, being found in the form of α-tocopherol and having a role in the prevention of various Email: [email protected] diseases, including: cancer, diabetes, cataracts and cardiovascular and cerebrovascular diseases. -
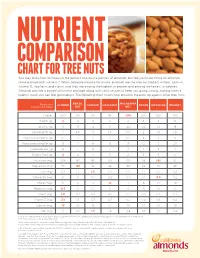
Chart for Tree Nuts
NUTRIENT COMPARISON CHART FOR TREE NUTS You may know how to measure the perfect one-ounce portion of almonds, but did you know those 23 almonds come packed with nutrients? When compared ounce for ounce, almonds are the tree nut highest in fiber, calcium, vitamin E, riboflavin and niacin, and they are among the highest in protein and among the lowest in calories. Almonds provide a powerful nutrient package along with tasty crunch to keep you going strong, making them a healthy snack you can feel good about. The following chart shows how almonds measure up against other tree nuts. BRAZIL MACADAMIA Based on a ALMOND CASHEW HAZELNUT PECAN PISTACHIO WALNUT one-ounce portion1 NUT NUT Calories 1602 190 160 180 200 200 160 190 Protein (g) 6 4 4 4 2 3 6 4 Total Fat (g) 14 19 13 17 22 20 13 18 Saturated Fat (g) 1 4.5 3 1.5 3.5 2 1.5 1.5 Polyunsaturated Fat (g) 3.5 7 2 2 0.5 6 4 13 Monounsaturated Fat (g) 9 7 8 13 17 12 7 2.5 Carbohydrates (g) 6 3 9 5 4 4 8 4 Dietary Fiber (g) 4 2 1 3 2 3 3 2 Potassium (mg) 208 187 160 193 103 116 285 125 Magnesium (mg) 77 107 74 46 33 34 31 45 Zinc (mg) 0.9 1.2 1.6 0.7 0.4 1.3 0.7 0.9 Vitamin B6 (mg) 0 0 0.1 0.2 0.1 0.1 0.3 0.2 Folate (mcg) 12 6 20 32 3 6 14 28 Riboflavin (mg) 0.3 0 0.1 0 0 0 0.1 0 Niacin (mg) 1.0 0.1 0.4 0.5 0.7 0.3 0.4 0.3 Vitamin E (mg) 7.3 1.6 0.3 4.3 0.2 0.4 0.6 0.2 Calcium (mg) 76 45 13 32 20 20 30 28 Iron (mg) 1.1 0.7 1.7 1.3 0.8 0.7 1.1 0.8 Source: U.S. -

Almond Cashew Barfi Recipe
150 Jackson St, Petone T: 568 4149 E: [email protected] ALMOND CASHEW BARFI RECIPE Almond Walnut Cashew Barfi is a healthy, quality substitute for candy that your entire family will enjoy Ingredients Makes 24 pcs 1/2 c almonds 1/2 c water 1/2 c walnuts 1/2 tsp cardamom powder 1/2 c cashew nuts 1 Tbsp sliced almonds to garnish 1 1/4 c sugar Method 1. Dry grind the walnuts, cashews and almonds in a food processor. 2. Dry roast the grounded nuts in a frying pan on low medium heat. 3. Roast them just enough so that the nuts start to give off an aroma. It will take about 4 to 5 minutes. Remove from heat and set aside. 4. Put the sugar and water together in a saucepan on medium heat. Bring to a boil to make the 1 thread syrup or on the candy thermometer it should reach 230 degrees F. 5. Turn off the heat and stir in the cardamom powder. 6. Add the nuts to the syrup and mix, and then spread over a greased 8-inch plate. Note: don’t let the syrup cool off. It must be spread while still hot. 7. Wait a few minutes until barfi is set but still soft. 8. Then cut the barfi into any shape you like (such as square, diamond, triangle). 9. Garnish each piece of barfi with sliced almonds while the barfi is still soft. 10. Allow the barfi to cool for about an hour to dry and hold its shape. -

Optimizing Almond Hulls for Dairy, Poultry and Insect Feedstocks in the U.S
OPTIMIZING ALMOND HULLS FOR DAIRY, POULTRY AND INSECT FEEDSTOCKS IN THE U.S. AND ABROAD ROOM 306-307 | DECEMBER 6, 2018 AGENDA Moderator: • Mike Curry, Johnson Farms Speakers: • Jed Asmus, January Innovation, Inc. • Woo Kyun Kim, University of Georgia • Jean VanderGheynst, UC Davis • Eric Tilton, HermetiaPro 2 Feedstuffs for Dairy Cow Diets: Considerations for Almond Hull use and Production Jed Asmus, M.S., PAS January Innovation Inc. What we are going to review: • The objective of Dairy producers, and how it relates to feeding cows • How feed stuffs are selected, measured and compared • Comparing Almond Hulls to other feeds commonly used • Information known about Almond Hulls • …. And the things we have yet to learn. • Concerns with feeding them. 4 The goal for Dairy Producers • Like all businesses, the objective is to be as profitable as possible. • If milk is money (which it is), the goal of all dairy producers is to produce milk as efficiently as possible. • The single largest cost center on a dairy is feed; Accounting for up to 60% of total expenses. • This means, that the more efficiently the cow uses the feed she consumes, the more productive she is, and can mean the more profitable she is. • Its worth remembering that milk is a commodity, with average profit margins running around 3-5% over time. 5 Which one to choose • Designing diets for dairy cattle is a very technical, and scientific undertaking – Variables considered when designing diets include Age of Cattle Cattle Grouping Milk production Risk Stage of Lactation Margin of Error Components Cash Flow Feed prices Genetics Feed quality Breed Feed Supply Geographical Location Weather Milk Pricing structure Labor Owners business objectives Equipment Future herd demographics 6 How the industry looks at feeds for diet design • Feed stuffs are compared on an analytical basis – Samples are sent out for chemical analysis, often resulting in 30+ variables being reported and used in diet design. -

Almond Pecan Corn
Almond Pecan Corn 2 cups pecan halves, lightly toasted 2 cups unblanched almonds, lightly toasted 8 cups plain popped popcorn 2 cups white sugar 1 cup golden corn syrup 2/3 cup water 2 teaspoons kosher salt 1 pound unsalted butter 1. Combine popcorn and nuts in a large bowl and toss to distribute nuts. Line 2 large baking sheets with foil or parchment paper and set aside. 2. Combine sugar, corn syrup and water in a large heavy saucepan (non- stick makes clean-up easier) and bring to a boil over high heat. Wash down any sugar crystals clinging to the sides of the pan with a pastry brush dipped in cold water. Clip a candy thermometer to the side of the pan, making sure that the tip does not touch the bottom of the pan. Add the salt and butter and continue cooking on high heat, without stirring, until candy thermometer registers 280-300º F. Do not leave the stove while this mixture is cooking. If it looks golden brown at 280º F remove from heat, if still pale, continue cooking to 300º F. 3. Wearing oven mitts carefully pour the hot syrup over the popcorn and nut mixture. Using two wooden spoons, sprayed with Pam, toss well to coat the mixture with the caramel. Spread the mixture into a thin layer on the foil lined baking sheets. Cool completely before breaking into bite size pieces. Store in an airtight container until ready to package for giving. This keeps well in an airtight container for about 2 weeks. -

A Technical Glance on Some Cosmetic Oils
European Scientific Journal June 2014 /SPECIAL/ edition vol.2 ISSN: 1857 – 7881 (Print) e - ISSN 1857- 7431 A TECHNICAL GLANCE ON SOME COSMETİC OİLS Kenan Yildirim, PhD Department of Fiber and Polymer Engineering Faculty of Natural Sciences, Architecture and Engineering,Bursa Technical University Bursa, Turkiye A. Melek Kostem, MSc Tübitak-Butal Bursa Test and Analysis Laboratory, Bursa Turkiye Abstract The properties of molecular structure, thermal behavior and UVA protection of 10 types of healthy promoting oils which are argania, almond, apricot seed, and jojoba, wheat germ, and sesame seed, avocado, cocoa, carrot and grapes seed oils were studied. FT-IR analysis was used for molecular structure. DSC analysis was used for thermal behavior and UV analysis was used for UV, visible and IR light absorption. Molecular structure and thermal behavior of avocado and cocoa are different from the others. Contrary to the spectra of avocado and cocoa on which the peaks belongs to carboxylic acid very strong, the spectra of the others do not involve carboxylic acid strong peaks. Almond, wheat germ and apricot seed oil absorb all UVA before 350 nm wave length. The UV protection property of grapes seed and cocoa is very well. Protection of wheat germ, almond and apricot seed oils to UVA is well respectively. Absorption of IR rate change from 7% to 25% for carrot, apricot seed, wheat germ, argania, avocado and grapes seed respectively. Keywords: Cosmetic oils, thermal behavior, molecular structure, UV protection Practical applications Except avocado and cocoa oils the others may be used for production of inherently UV protection yarn. This type of yarn can be used for producing UV protection clothes. -

Naturalization of Almond Trees (Prunus Dulcis) in Semi-Arid Regions
*Manuscript Click here to view linked References 1 2 3 4 5 6 7 8 9 Naturalization of almond trees (Prunus dulcis) in semi-arid 10 11 regions of the Western Mediterranean. 12 13 14 15 16 17 18 1 2 1,3 19 Pablo Homet-Gutierrez , Eugene W. Schupp , José M. Gómez * 20 1Departamento de Ecología, Facultad de Ciencias. Universidad de Granada, España. 21 2 22 Department of Wildland Resources and the Ecology Center. Utah State University. USA. 23 3Dpt de Ecología Evolutiva y Funcional, Estación Experimental de Zonas Aridas (EEZA-CSIC), 24 25 Almería, España. 26 27 28 *Corresponding author 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 1 63 64 65 Abstract 1 Agricultural land abandonment is rampant in present day Europe. A major consequence of this 2 3 phenomenon is the re-colonization of these areas by the original vegetation. However, some 4 agricultural, exotic species are able to naturalize and colonize these abandoned lands. In this 5 6 study we explore the ability of almonds (Prunus dulcis D.A. Webb.) to establish in abandoned 7 croplands in semi-arid areas of SE Iberian Peninsula. Domesticated during the early Holocene 8 9 in SW Asia and the Eastern Mediterranean, the almond has spread as a crop all over the world. 10 We established three plots adjacent to almond orchards on land that was abandoned and 11 12 reforested with Aleppo pine (Pinus halepensis Mill.) and Holm oak (Quercus ilex L.) about 20 13 years ago. -
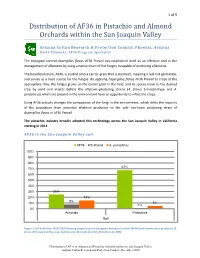
2020 Report on the Distribution of AF36 in Pistachio and Almond
1 of 5 Distribution of AF36 in Pistachio and Almond Orchards within the San Joaquin Valley Arizona Cotton Research & Protection Council, Phoenix, Arizona David Edmunds, AF36 Program Specialist The biological control Aspergillus flavus AF36 Prevail has established itself as an effective tool in the management of aflatoxins by using a native strain of the fungus incapable of producing aflatoxins. The beneficial strain, AF36, is coated onto a carrier grain that is sterilized, meaning it will not germinate, and serves as a food source for the fungus. By applying Aspergillus flavus AF36 Prevail to crops at the appropriate time, the fungus grows on the carrier grain in the field, and its spores move to the desired crop by wind and insects before the aflatoxin-producing strains (A. flavus S-morphotype and A. parasiticus) which are present in the environment have an opportunity to infect the crops. Using AF36 actually changes the composition of the fungi in the environment, which shifts the majority of the population from potential aflatoxin producers to the safe non-toxin producing strain of Aspergillus flavus in AF36 Prevail. The pistachio industry broadly adopted this technology across the San Joaquin Valley in California starting in 2012. AF36 in the San Joaquin Valley soil AF36 S-strains A. parasiticus 100% 90% 80% 67% 70% 60% 50% 40% 30% 25% 20% 14% 8% 10% 5% 1% 0% Almonds Pistachios Soil Figure 1 Soil data from 2018-2020 showing proportions of atoxigenic biological control (AF36) and known toxin-producers (S- strain, AP) separated by crop. Sample size: Almonds (n=237), Pistachios (n=390).