ACCEPTED VERSION Esmaeil Ebrahimie, Seyyed Hani Moussavi
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Multivariate Meta-Analysis of Differential Principal Components Underlying Human Primed and Naive-Like Pluripotent States
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.20.347666; this version posted October 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. This article is a US Government work. It is not subject to copyright under 17 USC 105 and is also made available for use under a CC0 license. October 20, 2020 To: bioRxiv Multivariate Meta-Analysis of Differential Principal Components underlying Human Primed and Naive-like Pluripotent States Kory R. Johnson1*, Barbara S. Mallon2, Yang C. Fann1, and Kevin G. Chen2*, 1Intramural IT and Bioinformatics Program, 2NIH Stem Cell Unit, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, Maryland 20892, USA Keywords: human pluripotent stem cells; naive pluripotency, meta-analysis, principal component analysis, t-SNE, consensus clustering *Correspondence to: Dr. Kory R. Johnson ([email protected]) Dr. Kevin G. Chen ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.10.20.347666; this version posted October 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. This article is a US Government work. It is not subject to copyright under 17 USC 105 and is also made available for use under a CC0 license. ABSTRACT The ground or naive pluripotent state of human pluripotent stem cells (hPSCs), which was initially established in mouse embryonic stem cells (mESCs), is an emerging and tentative concept. To verify this important concept in hPSCs, we performed a multivariate meta-analysis of major hPSC datasets via the combined analytic powers of percentile normalization, principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), and SC3 consensus clustering. -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/) -

Proteomic Analysis Uncovers Measles Virus Protein C Interaction with P65
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.08.084418; this version posted May 9, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Proteomic Analysis Uncovers Measles Virus Protein C Interaction with p65/iASPP/p53 Protein Complex Alice Meignié1,2*, Chantal Combredet1*, Marc Santolini 3,4, István A. Kovács4,5,6, Thibaut Douché7, Quentin Giai Gianetto 7,8, Hyeju Eun9, Mariette Matondo7, Yves Jacob10, Regis Grailhe9, Frédéric Tangy1**, and Anastassia V. Komarova1, 10** 1 Viral Genomics and Vaccination Unit, Department of Virology, Institut Pasteur, CNRS UMR-3569, 75015 Paris, France 2 Université Paris Diderot, Sorbonne Paris Cité, Paris, France 3 Center for Research and Interdisciplinarity (CRI), Université de Paris, INSERM U1284 4 Network Science Institute and Department of Physics, Northeastern University, Boston, MA 02115, USA 5 Department of Physics and Astronomy, Northwestern University, Evanston, IL 60208-3109, USA 6 Department of Network and Data Science, Central European University, Budapest, H-1051, Hungary 7 Proteomics platform, Mass Spectrometry for Biology Unit (MSBio), Institut Pasteur, CNRS USR 2000, Paris, France. 8 Bioinformatics and Biostatistics Hub, Computational Biology Department, Institut Pasteur, CNRS USR3756, Paris, France 9 Technology Development Platform, Institut Pasteur Korea, Seongnam-si, Republic of Korea 10 Laboratory of Molecular Genetics of RNA Viruses, Institut Pasteur, CNRS UMR-3569, -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
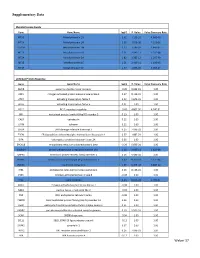
Weber 37 Supplementary Data
Supplementary Data Metallothionein Family Gene Gene Name logFC P. Value False Discovery Rate MT1G Metallothionein 1G 2.62 1.19E-10 4.66E-08 MT1H Metallothionein 1H 2.58 4.87E-08 7.12E-06 MT1M Metallothionein 1M 2.23 1.18E-07 1.44E-05 MT1X Metallothionein 1X 2.21 2.64E-11 1.25E-08 MT2A Metallothionein 2A 1.82 1.38E-12 1.26E-09 MT1E Metallothionein 1E 1.51 2.41E-11 1.18E-08 MT1F Metallothionein 1F 1.47 1.85E-09 4.09E-07 Unfolded Protein Response Gene Gene Name logFC P. Value False Discovery Rate AMFR autocrine motility factor receptor -0.20 9.94E-01 1.00 ASK1 mitogen-activated protein kinase kinase kinase 5 0.27 8.11E-01 1.00 ATF4 activating transcription factor 4 0.22 9.67E-01 1.00 ATF6 activating transcription factor 6 0.01 1.00 1.00 BCL2 BCL2, apoptosis regulator -0.43 4.80E-02 4.24E-01 BIP heat shock protein family A (Hsp70) member 5 0.13 1.00 1.00 CALR calreticulin 0.12 1.00 1.00 CANX calnexin 0.12 1.00 1.00 CHOP DNA damage inducible transcript 3 0.25 7.99E-01 1.00 EDEM ER degradation enhancing alpha-mannosidase like protein 1 0.37 1.88E-01 1.00 EIFA eukaryotic translation initiation factor 2A 0.06 1.00 1.00 ERO1LB endoplasmic reticulum oxidoreductase 1 beta -0.29 5.69E-01 1.00 GADD34 protein phosphatase 1 regulatory subunit 15A 1.18 3.66E-11 1.62E-08 GRP94 heat shock protein 90 beta family member 1 0.16 9.98E-01 1.00 HSPH1 heat shock protein family H (Hsp110) member 1 0.62 9.70E-06 5.12E-04 INSIG1 insulin induced gene 1 1.69 8.47E-18 3.88E-14 IRE1 endoplasmic reticulum to nucleus signaling 1 0.26 8.13E-01 1.00 JNK1 mitogen-activated -

Identification of Potential Key Genes and Pathway Linked with Sporadic Creutzfeldt-Jakob Disease Based on Integrated Bioinformatics Analyses
medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Identification of potential key genes and pathway linked with sporadic Creutzfeldt-Jakob disease based on integrated bioinformatics analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Abstract Sporadic Creutzfeldt-Jakob disease (sCJD) is neurodegenerative disease also called prion disease linked with poor prognosis. The aim of the current study was to illuminate the underlying molecular mechanisms of sCJD. The mRNA microarray dataset GSE124571 was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were screened. -

Supplement 1A Steffensen Et
Liver Wild-type Knockout C T C T 1 2 4 7 8 9 1 2 4 5 7 9 1 1 1 1 1 1 C C C T T T C C C T T T W W W W W W K K K K K K IMAGE:793166 RIKEN cDNA 6720463E02 gene IMAGE:1447421 ESTs, Weakly similar to ZF37 MOUSE ZINC FINGER PROTEIN 37 [M.musculus] IMAGE:934291 RIKEN cDNA 2810418N01 gene IMAGE:1247525 small EDRK-rich f2actor IMAGE:1449402 expressed sequence AW321064 IMAGE:1279847 ESTs IMAGE:518737 expressed sequence AW049941 IMAGE:860231 a disintegrin and metalloproteinase domain 17 IMAGE:642836 CD86 antigen IMAGE:1003885 phosphoribosyl pyrophosphate sy1nthetase IMAGE:524862 RIKEN cDNA 5730469D23 gene IMAGE:1264473 protein inhibitor of activat1ed STAT IMAGE:847035 RIKEN cDNA 4833422F06 gene IMAGE:374550 requiem IMAGE:976520 nuclear receptor coact4ivator IMAGE:1264311 Unknown IMAGE:976735 expressed sequence AI987692 IMAGE:976659 cathepsLin IMAGE:1477580 RIKEN cDNA 1600010J02 gene IMAGE:1277168 ribosomal protein, large, P1 IMAGE:524842 RIKEN cDNA 0710008D09 gene IMAGE:373019 split hand/foot delete1d gene IMAGE:404428 expressed sequence AI413851 IMAGE:619810 RIKEN cDNA 1700003F10 gene IMAGE:1749558 caspase 3, apoptosis related cysteine protease IMAGE:718718 RIKEN cDNA 2810003F23 gene IMAGE:819789 Unknown IMAGE:524474 ATP-binding cassette, sub-family A ABC1, member IMAGE:804950 Mus musculus, Similar to ribosomal protein S20, clone MGC:6876 IMAGE:2651405, mRNA, complete cds IMAGE:806143 gap junction membrane channel prot2ein beta IMAGE:1745887 expressed sequence AI836376 IMAGE:779426 RIKEN cDNA 5230400G24 gene IMAGE:1125615 Unknown IMAGE:535025 DNA -

Literature Mining Sustains and Enhances Knowledge Discovery from Omic Studies
LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES by Rick Matthew Jordan B.S. Biology, University of Pittsburgh, 1996 M.S. Molecular Biology/Biotechnology, East Carolina University, 2001 M.S. Biomedical Informatics, University of Pittsburgh, 2005 Submitted to the Graduate Faculty of School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2016 UNIVERSITY OF PITTSBURGH SCHOOL OF MEDICINE This dissertation was presented by Rick Matthew Jordan It was defended on December 2, 2015 and approved by Shyam Visweswaran, M.D., Ph.D., Associate Professor Rebecca Jacobson, M.D., M.S., Professor Songjian Lu, Ph.D., Assistant Professor Dissertation Advisor: Vanathi Gopalakrishnan, Ph.D., Associate Professor ii Copyright © by Rick Matthew Jordan 2016 iii LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES Rick Matthew Jordan, M.S. University of Pittsburgh, 2016 Genomic, proteomic and other experimentally generated data from studies of biological systems aiming to discover disease biomarkers are currently analyzed without sufficient supporting evidence from the literature due to complexities associated with automated processing. Extracting prior knowledge about markers associated with biological sample types and disease states from the literature is tedious, and little research has been performed to understand how to use this knowledge to inform the generation of classification models from ‘omic’ data. Using pathway analysis methods to better understand the underlying biology of complex diseases such as breast and lung cancers is state-of-the-art. However, the problem of how to combine literature- mining evidence with pathway analysis evidence is an open problem in biomedical informatics research. -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

The Use of Phosphoproteomic Data to Identify Altered Kinases and Signaling Pathways in Cancer
The use of phosphoproteomic data to identify altered kinases and signaling pathways in cancer By Sara Renee Savage Thesis Submitted to the Faculty of the Graduate School of Vanderbilt University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE in Biomedical Informatics August 10, 2018 Nashville, Tennessee Approved: Bing Zhang, Ph.D. Carlos Lopez, Ph.D. Qi Liu, Ph.D. ACKNOWLEDGEMENTS The work presented in this thesis would not have been possible without the funding provided by the NLM training grant (T15-LM007450) and the support of the Biomedical Informatics department at Vanderbilt. I am particularly indebted to Rischelle Jenkins, who helped me solve all administrative issues. Furthermore, this work is the result of a collaboration between all members of the Zhang lab and the larger CPTAC consortium. I would like to thank the other CPTAC centers for processing the data, and Chen Huang and Suhas Vasaikar in the Zhang lab for analyzing the colon cancer copy number and proteomic data, respectively. All members of the Zhang lab have been extremely helpful in answering any questions I had and offering suggestions on my work. Finally, I would like to acknowledge my mentor, Bing Zhang. I am extremely grateful for his guidance and for giving me the opportunity to work on these projects. ii TABLE OF CONTENTS Page ACKNOWLEDGEMENTS ................................................................................................ ii LIST OF TABLES............................................................................................................