Yeasts and Yeast-Like Fungi As an Element of Purity Assessment of Surface Waters
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Vles) in the Yeast Debaryomyces Hansenii
toxins Article New Cytoplasmic Virus-Like Elements (VLEs) in the Yeast Debaryomyces hansenii Xymena Połomska 1,* ,Cécile Neuvéglise 2, Joanna Zyzak 3, Barbara Zarowska˙ 1, Serge Casaregola 4 and Zbigniew Lazar 1 1 Department of Biotechnology and Food Microbiology, Faculty of Biotechnology and Food Science, Wrocław University of Environmental and Life Sciences (WUELS), 50-375 Wroclaw, Poland; [email protected] (B.Z.);˙ [email protected] (Z.L.) 2 SPO, INRAE, Montpellier SupAgro, Université de Montpellier, 34060 Montpellier, France; [email protected] 3 Department of Microbiology, Laboratory of Microbiome Immunobiology, Ludwik Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, 53-114 Wroclaw, Poland; [email protected] 4 INRAE, AgroParisTech, Micalis Institute, CIRM-Levures, Université Paris-Saclay, 78350 Jouy-en-Josas, France; [email protected] * Correspondence: [email protected]; Tel.: +48-71-3207-791 Abstract: Yeasts can have additional genetic information in the form of cytoplasmic linear dsDNA molecules called virus-like elements (VLEs). Some of them encode killer toxins. The aim of this work was to investigate the prevalence of such elements in D. hansenii killer yeast deposited in culture collections as well as in strains freshly isolated from blue cheeses. Possible benefits to the host from harboring such VLEs were analyzed. VLEs occurred frequently among fresh D. hansenii isolates (15/60 strains), as opposed to strains obtained from culture collections (0/75 strains). Eight new different systems were identified: four composed of two elements and four of three elements. Full sequences of three new VLE systems obtained by NGS revealed extremely high conservation Citation: Połomska, X.; Neuvéglise, among the largest molecules in these systems except for one ORF, probably encoding a protein C.; Zyzak, J.; Zarowska,˙ B.; resembling immunity determinant to killer toxins of VLE origin in other yeast species. -

Comparative Genomics of Protoploid Saccharomycetaceae
Downloaded from genome.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Evolution of protoploid yeast genomes ___________________________________________________________________________ Comparative genomics of protoploid Saccharomycetaceae. The Génolevures Consortium (1) Running title: Evolution of protoploid yeast genomes Key words: protein families, synteny, tandems, annotation, SONS, ancestor genome Corresponding author: Jean Luc Souciet Université de Strasbourg, CNRS, UMR 7156 Institut de Botanique, 28 rue Goethe, F-67000 Strasbourg, France Tel: 33 3 90 24 18 17 FAX: 33 3 90 24 20 28 e-mail: [email protected] (1) List of participants and affiliations appear at the end of the paper 1 Downloaded from genome.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Evolution of protoploid yeast genomes ___________________________________________________________________________ Abstract Our knowledge on yeast genomes remains largely dominated by the extensive studies on Saccharomyces cerevisiae and the consequences of its ancestral duplication, leaving the evolution of the entire class of hemiascomycetes only partly explored. We concentrate here on five species of Saccharomycetaceae, a large subdivision of hemiascomycetes, that we call “protoploid” because they diverged from the S. cerevisiae lineage prior to its genome duplication. We determined the complete genome sequences of three of these species, Kluyveromyces (Lachancea) thermotolerans and Saccharomyces (Lachancea) kluyveri (two members of the newly described Lachancea clade) and Zygosaccharomyces rouxii. We included in our comparisons the previously available sequences of Klyveromyces lactis and Ashbya (Eremothecium) gossypii. Despite their broad evolutionary range and significant individual variations in each lineage, the five protoploid Saccharomycetaceae share a core repertoire of ca. -
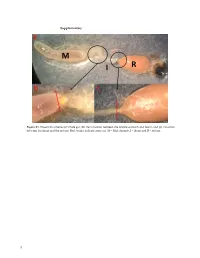
Supplementary File 1
Supplementary Figure S1. Dissection scheme (a) whole gut, (b) the transition between the middle stomach and ileum, and (c) transition between the ileum and the rectum. Red streaks indicate areas cut. M = Mid stomach, I = ileum and R = rectum. 1 Figure S2. Relative abundance of the 20 most abundant fungal OTUs across months per gut part. Taxonomic assignments are provided in Table S1. 2 Figure S3. Relative abundance of the bacterial OUTs (with abundance > 1%) across months per gut part. The color code is given at the genus level. 3 Table S1. Taxonomic assignments of fungal OTUs by Blast. OTU# Taxonomy by blast nt collection 11/11‐2020 Phylum Accesion# OTU_1 Aureobasidium pullulans Ascomycota MW085051 OTU_2 Meyerozyma guilliermondii Ascomycota MT988167 OTU_3 Starmerella apicola Ascomycota KY101940 OTU_4 Debaryomyces hansenii Ascomycota MW051606 OTU_5 Engyodontium sp. Ascomycota MN905797 Basidiomy‐ OTU_6 Mrakia sp. MT505696 cota Basidiomy‐ OTU_7 Tausonia pullulans MN900123 cota OTU_8 Unknown OTU_9 Unknown OTU_10 Penicillium corylophilum Ascomycota MT906500 OTU_11 Penicillium sp. Ascomycota MT993349 OTU_12 Cladosporium sp. Ascomycota MW077705 Phaffia rhodozyma/Xanthophyllomyces dendro‐ Basidiomy‐ OTU_13 KY104501/DQ904243 rhous cota Basidiomy‐ OTU_14 Filobasidium wieringae MN899199 cota OTU_15 Hanseniaspora uvarum Ascomycota MN556596 Basidiomy‐ OTU_16 Mrakia gelida MN460370 cota OTU_17 Uncultured fungus clone MK717974 OTU_18 Cladosporium allicinum Ascomycota MT974153 OTU_20 Taphrina carpini Ascomycota MK782181 Basidiomy‐ OTU_25 Papiliotrema -

High-Level Classification of the Fungi and a Tool for Evolutionary Ecological Analyses
Fungal Diversity (2018) 90:135–159 https://doi.org/10.1007/s13225-018-0401-0 (0123456789().,-volV)(0123456789().,-volV) High-level classification of the Fungi and a tool for evolutionary ecological analyses 1,2,3 4 1,2 3,5 Leho Tedersoo • Santiago Sa´nchez-Ramı´rez • Urmas Ko˜ ljalg • Mohammad Bahram • 6 6,7 8 5 1 Markus Do¨ ring • Dmitry Schigel • Tom May • Martin Ryberg • Kessy Abarenkov Received: 22 February 2018 / Accepted: 1 May 2018 / Published online: 16 May 2018 Ó The Author(s) 2018 Abstract High-throughput sequencing studies generate vast amounts of taxonomic data. Evolutionary ecological hypotheses of the recovered taxa and Species Hypotheses are difficult to test due to problems with alignments and the lack of a phylogenetic backbone. We propose an updated phylum- and class-level fungal classification accounting for monophyly and divergence time so that the main taxonomic ranks are more informative. Based on phylogenies and divergence time estimates, we adopt phylum rank to Aphelidiomycota, Basidiobolomycota, Calcarisporiellomycota, Glomeromycota, Entomoph- thoromycota, Entorrhizomycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota and Olpidiomycota. We accept nine subkingdoms to accommodate these 18 phyla. We consider the kingdom Nucleariae (phyla Nuclearida and Fonticulida) as a sister group to the Fungi. We also introduce a perl script and a newick-formatted classification backbone for assigning Species Hypotheses into a hierarchical taxonomic framework, using this or any other classification system. We provide an example -

The 100 Years of the Fungus Collection Mucl 1894-1994
THE 100 YEARS OF THE FUNGUS COLLECTION MUCL 1894-1994 Fungal Taxonomy and Tropical Mycology: Quo vadis ? Taxonomy and Nomenclature of the Fungi Grégoire L. Hennebert Catholic University of Louvain, Belgium Notice of the editor This document is now published as an archive It is available on www.Mycotaxon.com It is also produced on CD and in few paperback copies G. L. Hennebert ed. Published by Mycotaxon, Ltd. Ithaca, New York, USA December 2010 ISBN 978-0-930845-18-6 (www pdf version) ISBN 978-0-930845-17-9 (paperback version) DOI 10.5248/2010MUCL.pdf 1894-1994 MUCL Centenary CONTENTS Lists of participants 8 Forword John Webser 13 PLENARY SESSION The 100 Year Fungus Culture Collection MUCL, June 29th, 1994 G.L. Hennebert, UCL Mycothèque de l'Université Catholique de Louvain (MUCL) 17 D. Hawksworth, IMI, U.K. Fungal genetic resource collections and biodiversity. 27 D. van der Mei, CBS, MINE, Netherlands The fungus culture collections in Europe. 34 J. De Brabandere, BCCM, Belgium The Belgian Coordinated Collections of Microorganisms. 40 Fungal Taxonomy and tropical Mycology G.L. Hennebert, UCL Introduction. Fungal taxonomy and tropical mycology: Quo vadis ? 41 C.P. Kurtzman, NRRL, USA Molecular taxonomy in the yeast fungi: present and future. 42 M. Blackwell, Louisiana State University, USA Phylogeny of filamentous fungi deduced from analysis of molecular characters: present and future. 52 J. Rammeloo, National Botanical Garden, Belgium Importance of morphological and anatomical characters in fungal taxonomy. 57 M.F. Roquebert, Natural History Museum, France Possible progress of modern morphological analysis in fungal taxonomy. 63 A.J. -

Diversity and Distribution of Hidden Cultivable Fungi Associated with Marine Animals of Antarctica Fungal Biology
Fungal Biology 123 (2019) 507e516 Contents lists available at ScienceDirect Fungal Biology journal homepage: www.elsevier.com/locate/funbio Diversity and distribution of hidden cultivable fungi associated with marine animals of Antarctica Valeria Martins Godinho a, Maria Theresa Rafaela de Paula a, Debora Amorim Saraiva Silva a, Karla Paresque b, Aline Paternostro Martins c, * Pio Colepicolo c, Carlos Augusto Rosa a, Luiz Henrique Rosa a, a Departamento de Microbiologia, Instituto de Ci^encias Biologicas, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil b Laboratorio de Bentologia, Universidade Federal de Alagoas, Maceio, AL, Brazil c Departamento de Bioquímica, Instituto de Química, Universidade de Sao~ Paulo, Sao~ Paulo, Brazil article info abstract Article history: In the present study, we surveyed the distribution and diversity of fungal assemblages associated with 10 Received 22 November 2018 species of marine animals from Antarctica. The collections yielded 83 taxa from 27 distinct genera, which Received in revised form were identified using molecular biology methods. The most abundant taxa were Cladosporium sp. 1, 17 April 2019 Debaryomyces hansenii, Glaciozyma martinii, Metschnikowia australis, Pseudogymnoascus destructans, Accepted 1 May 2019 Thelebolus cf. globosus, Pseudogymnoascus pannorum, Tolypocladium tundrense, Metschnikowia australis, Available online 14 May 2019 and different Penicillium species. The diversity, richness, and dominance of fungal assemblages ranged Corresponding Editor: Nik Money among the host; however, in general, the fungal community, which was composed of endemic and cold- adapted cosmopolitan taxa distributed across the different sites of Antarctic Peninsula, displayed high Keywords: diversity, richness, and dominance indices. Our results contribute to knowledge about fungal diversity in Antarctic ocean the marine environment across the Antarctic Peninsula and their phylogenetic relationships with species Extremophiles that occur in other cold, temperate, and tropical regions of the World. -

Descriptions of Medical Fungi
DESCRIPTIONS OF MEDICAL FUNGI THIRD EDITION (revised November 2016) SARAH KIDD1,3, CATRIONA HALLIDAY2, HELEN ALEXIOU1 and DAVID ELLIS1,3 1NaTIONal MycOlOgy REfERENcE cENTRE Sa PaTHOlOgy, aDElaIDE, SOUTH aUSTRalIa 2clINIcal MycOlOgy REfERENcE labORatory cENTRE fOR INfEcTIOUS DISEaSES aND MIcRObIOlOgy labORatory SERvIcES, PaTHOlOgy WEST, IcPMR, WESTMEaD HOSPITal, WESTMEaD, NEW SOUTH WalES 3 DEPaRTMENT Of MOlEcUlaR & cEllUlaR bIOlOgy ScHOOl Of bIOlOgIcal ScIENcES UNIvERSITy Of aDElaIDE, aDElaIDE aUSTRalIa 2016 We thank Pfizera ustralia for an unrestricted educational grant to the australian and New Zealand Mycology Interest group to cover the cost of the printing. Published by the authors contact: Dr. Sarah E. Kidd Head, National Mycology Reference centre Microbiology & Infectious Diseases Sa Pathology frome Rd, adelaide, Sa 5000 Email: [email protected] Phone: (08) 8222 3571 fax: (08) 8222 3543 www.mycology.adelaide.edu.au © copyright 2016 The National Library of Australia Cataloguing-in-Publication entry: creator: Kidd, Sarah, author. Title: Descriptions of medical fungi / Sarah Kidd, catriona Halliday, Helen alexiou, David Ellis. Edition: Third edition. ISbN: 9780646951294 (paperback). Notes: Includes bibliographical references and index. Subjects: fungi--Indexes. Mycology--Indexes. Other creators/contributors: Halliday, catriona l., author. Alexiou, Helen, author. Ellis, David (David H.), author. Dewey Number: 579.5 Printed in adelaide by Newstyle Printing 41 Manchester Street Mile End, South australia 5031 front cover: Cryptococcus neoformans, and montages including Syncephalastrum, Scedosporium, Aspergillus, Rhizopus, Microsporum, Purpureocillium, Paecilomyces and Trichophyton. back cover: the colours of Trichophyton spp. Descriptions of Medical Fungi iii PREFACE The first edition of this book entitled Descriptions of Medical QaP fungi was published in 1992 by David Ellis, Steve Davis, Helen alexiou, Tania Pfeiffer and Zabeta Manatakis. -

Human Gut Mycobiome in Ibd | Encyclopedia
Human Gut Mycobiome in IBD Created by: Mario Matijasic , Tomislav Meštrović , Hana Čipčić Paljetak , Mihaela Perić , Anja Barešić , Donatella Verbanac Version received: 16 April 2020 The human microbiota is a diverse microbial ecosystem associated with many beneficial physiological functions, as well as numerous disease etiologies. Dominated by bacteria, the microbiota also includes commensal populations of fungi, viruses, archaea, and protists. Unlike bacterial microbiota, which was extensively studied in the past two decades, these non-bacterial microorganisms, their functional roles, and their interaction with one another or with host immune system have not been as widely explored. This review covers the recent findings on the fungal communities of the human gastrointestinal microbiota, termed the “mycobiome”, and their involvement in health and disease, with particular focus on the pathophysiology of inflammatory bowel disease. Introduction Fungi are ubiquitous in the environment and a part of all Earth’s ecosystems[1]. In addition, a diverse population of commensal fungi has been recognized as a fundamental component of the human body, co-existing with other microbes within the human microbiota.[2] In contrast to the vast number of studies on the bacterial communities of the microbiota conducted in the last decades, the fungal constituents of the microbiota, the mycobiome, received much less attention. Still, recent research acknowledged human mycobiome as a dynamic community, responsive to environmental and pathophysiological changes, and playing a vital role in host metabolism as well as in maintenance of host immune homeostasis.[2][3][4][5] Human mycobiome is also implicated in various disease conditions, including inflammatory bowel disease (IBD) and its two main entities: Crohn’s disease (CD) and ulcerative colitis (UC). -
Comparative Genomics of Protoploid Saccharomycetaceae
Downloaded from genome.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press Article Comparative genomics of protoploid Saccharomycetaceae The Ge´nolevures Consortium1 Our knowledge of yeast genomes remains largely dominated by the extensive studies on Saccharomyces cerevisiae and the consequences of its ancestral duplication, leaving the evolution of the entire class of hemiascomycetes only partly ex- plored. We concentrate here on five species of Saccharomycetaceae, a large subdivision of hemiascomycetes, that we call ‘‘protoploid’’ because they diverged from the S. cerevisiae lineage prior to its genome duplication. We determined the complete genome sequences of three of these species: Kluyveromyces (Lachancea) thermotolerans and Saccharomyces (Lachancea) kluyveri (two members of the newly described Lachancea clade), and Zygosaccharomyces rouxii. We included in our comparisons the previously available sequences of Kluyveromyces lactis and Ashbya (Eremothecium) gossypii. Despite their broad evolutionary range and significant individual variations in each lineage, the five protoploid Saccharomycetaceae share a core repertoire of approximately 3300 protein families and a high degree of conserved synteny. Synteny blocks were used to define gene orthology and to infer ancestors. Far from representing minimal genomes without redundancy, the five protoploid yeasts contain numerous copies of paralogous genes, either dispersed or in tandem arrays, that, altogether, constitute a third of each genome. Ancient, conserved paralogs as well as novel, lineage-specific paralogs were identified. [Supplemental material is available online at http://www.genome.organd at http://www.genolevures.org/.The sequence data for Zygosaccharomyces rouxii and Kluyveromyces thermotolerans have been submitted to EMBL-Bank (http://www.ebi.ac.uk/ embl/) under accession nos. -
Comparative Genomics of Biotechnologically Important Yeasts
Comparative genomics of biotechnologically important yeasts Robert Rileya, Sajeet Haridasa, Kenneth H. Wolfeb, Mariana R. Lopesc,d, Chris Todd Hittingerc,e, Markus Gökerf, Asaf A. Salamova, Jennifer H. Wisecaverg, Tanya M. Longh,i, Christopher H. Calveyj, Andrea L. Aertsa, Kerrie W. Barrya, Cindy Choia, Alicia Cluma, Aisling Y. Coughlanb, Shweta Deshpandea, Alexander P. Douglassb, Sara J. Hansonb, Hans-Peter Klenkf,k, Kurt M. LaButtia, Alla Lapidusa,1, Erika A. Lindquista, Anna M. Lipzena, Jan P. Meier-Kolthofff, Robin A. Ohma,2, Robert P. Otillara, Jasmyn L. Pangilinana, Yi Penga, Antonis Rokasg, Carlos A. Rosad, Carmen Scheunerf, Andriy A. Sibirnyl,m, Jason C. Slotn, J. Benjamin Stielowf,o, Hui Suna, Cletus P. Kurtzmanp, Meredith Blackwellq,r, Igor V. Grigorieva,3, and Thomas W. Jeffriesh,3 aDepartment of Energy Joint Genome Institute, Walnut Creek, CA 94598; bUniversity College Dublin Conway Institute, School of Medicine, University College Dublin, Dublin 4, Ireland; cLaboratory of Genetics, Genetics/Biotechnology Center, University of Wisconsin–Madison, Madison, WI 53706; dDepartamento de Microbiologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, MG, 31270-901, Brazil; eDepartment of Energy Great Lakes Bioenergy Research Center, University of Wisconsin–Madison, Madison, WI 53726; fDeutsche Sammlung von Mikroorganismen und Zellkulturen German Collection of Microorganisms and Cell Cultures, Leibniz Institute, 38124 Braunschweig, Germany; gDepartment of Biological Sciences, Vanderbilt University, -

Diversity, Distribution, and Ecology of Fungi in the Seasonal Snow of Antarctica
microorganisms Article Diversity, Distribution, and Ecology of Fungi in the Seasonal Snow of Antarctica Graciéle C.A. de Menezes 1, Soraya S. Amorim 1,Vívian N. Gonçalves 1, Valéria M. Godinho 1, Jefferson C. Simões 2, Carlos A. Rosa 1 and Luiz H. Rosa 1,* 1 Departamento de Microbiologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte 31270-901, Brazil; [email protected] (G.C.A.d.M.); [email protected] (S.S.A.); [email protected] (V.N.G.); [email protected] (V.M.G.); [email protected] (C.A.R.) 2 Centro Polar e Climático, Universidade Federal do Rio Grande do Sul, Porto Alegre 91201-970, Brazil; jeff[email protected] * Correspondence: [email protected]; Tel.: +55-31-3409-2749 Received: 13 September 2019; Accepted: 8 October 2019; Published: 12 October 2019 Abstract: We characterized the fungal community found in the winter seasonal snow of the Antarctic Peninsula. From the samples of snow, 234 fungal isolates were obtained and could be assigned to 51 taxa of 26 genera. Eleven yeast species displayed the highest densities; among them, Phenoliferia glacialis showed a broad distribution and was detected at all sites that were sampled. Fungi known to be opportunistic in humans were subjected to antifungal minimal inhibition concentration. Debaryomyces hansenii, Rhodotorula mucilaginosa, Penicillium chrysogenum, Penicillium sp. 3, and Penicillium sp. 4 displayed resistance against the antifungals benomyl and fluconazole. Among them, R. mucilaginosa isolates were able to grow at 37 ◦C. Our results show that the winter seasonal snow of the Antarctic Peninsula contains a diverse fungal community dominated by cosmopolitan ubiquitous fungal species previously found in tropical, temperate, and polar ecosystems. -

An Online Resource for Marine Fungi
Fungal Diversity https://doi.org/10.1007/s13225-019-00426-5 (0123456789().,-volV)(0123456789().,- volV) An online resource for marine fungi 1,2 3 1,4 5 6 E. B. Gareth Jones • Ka-Lai Pang • Mohamed A. Abdel-Wahab • Bettina Scholz • Kevin D. Hyde • 7,8 9 10 11 12 Teun Boekhout • Rainer Ebel • Mostafa E. Rateb • Linda Henderson • Jariya Sakayaroj • 13 6 6,17 14 Satinee Suetrong • Monika C. Dayarathne • Vinit Kumar • Seshagiri Raghukumar • 15 1 16 6 K. R. Sridhar • Ali H. A. Bahkali • Frank H. Gleason • Chada Norphanphoun Received: 3 January 2019 / Accepted: 20 April 2019 Ó School of Science 2019 Abstract Index Fungorum, Species Fungorum and MycoBank are the key fungal nomenclature and taxonomic databases that can be sourced to find taxonomic details concerning fungi, while DNA sequence data can be sourced from the NCBI, EBI and UNITE databases. Nomenclature and ecological data on freshwater fungi can be accessed on http://fungi.life.illinois.edu/, while http://www.marinespecies.org/provides a comprehensive list of names of marine organisms, including information on their synonymy. Previous websites however have little information on marine fungi and their ecology, beside articles that deal with marine fungi, especially those published in the nineteenth and early twentieth centuries may not be accessible to those working in third world countries. To address this problem, a new website www.marinefungi.org was set up and is introduced in this paper. This website provides a search facility to genera of marine fungi, full species descriptions, key to species and illustrations, an up to date classification of all recorded marine fungi which includes all fungal groups (Ascomycota, Basidiomycota, Blastocladiomycota, Chytridiomycota, Mucoromycota and fungus-like organisms e.g.